Abstract
Mechanotransduction, the interplay between physical and chemical signaling, plays vital roles in many biological processes ranging from cell differentiation to metastasis. The state-of-the-art techniques to quantify cell forces employ deformable polymer films or molecular probes tethered to glass substrates. These types of flat substrates limit applications in investigating mechanotransduction on non-planar geometries where physiological activities such as phagocytosis and immunological synapse formation mostly occur. A second challenge is the low throughput of microscopy readout which limits the application of current assays in fundamental and clinical research. We address these challenges by developing a DNA-based microparticle tension sensor (μTS), which features a spherical surface and thus allows for investigation of mechanical events at curved interfaces or within groups of cells in suspension. Importantly, the micron-scale of μTS enables flow cytometry readout, which is rapid and high throughput. To demonstrate the scope of μTS, we applied the method to map and measure T-cell receptor (TCR) forces and platelet integrin forces at 12 and 56 pN thresholds. Furthermore, we quantified the inhibition efficiency of two anti-platelet drugs providing a proof-of-concept demonstration of μTS to screen drugs that modulate cellular mechanics.
Keywords: molecular tension sensor, drug screening, high-throughput quantification, non-planar geometry
Graphical Abstract
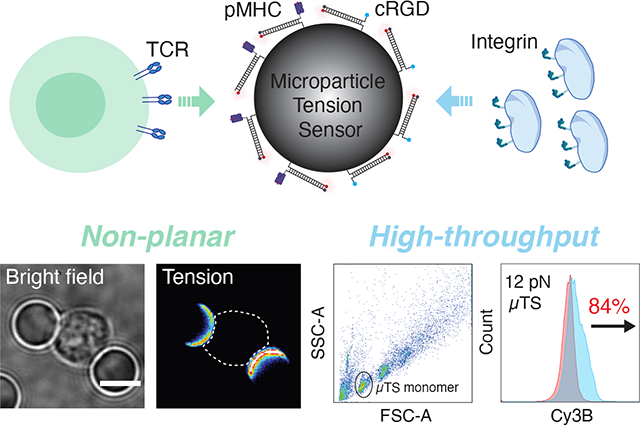
A microparticle tension sensor is designed for measuring molecular forces in non-planar geometries and for high-throughput flow cytometry analysis of cell mechanics.
Introduction
Cells function as force generators and sensors, constantly transmitting molecular forces to their extracellular matrix ligands and adjacent cells. These mechanical forces are generated by actomyosin contractility and actin polymerization and play vital roles in numerous activities throughout the lifetime of a cell including adhesion, migration and differentiation.[1] Mechanotransduction, or the coupling between mechanical forces and biochemical signaling, is mediated at the molecular level through individual receptor-ligand bonds.[2] To date, many strategies have been developed to measure cell forces. Amongst them, traction force microscopy (TFM) maps force at the single cell and subcellular length scale by tracking deformation of elastic substrates underneath cells.[3] However, the length scale and force range of these methods are limited to μm and nN respectively, thus hindering high resolution imaging and investigation of mechanotransduction at the molecular (pN) level. To measure the receptor forces applied to individual ligands, our lab previously developed molecular tension fluorescence microscopy (MTFM), in which “spring-like” extendable molecules, such as polymers of PEG with defined lengths,[4] proteins,[5] and DNA hairpin,[6] are engineered as fluorescence force sensors and generate turn-on fluorescence signal in response to molecular forces transmitted by cells through their membrane receptors. A complementary method to MTFM that was originally developed to manipulate cell forces, and then later adapted to recording cell force history is the DNA-based tension gauge tether (TGT) which was developed by Ha and colleagues.[7] In this method, a DNA duplex is irreversibly denatured when it experiences forces that exceed the tension tolerance (Ttol), causing a change in fluorescence signal. Unlike thermal or chemical denaturation, the mechanical energy that leads to DNA denaturation is highly dependent on force orientation. For example, the Ttol value is maximum (~60 pN) when a duplex experiences forces in a shearing geometry, with forces parallel to the long axis of the duplex; in contrast, forces perpendicular to the duplex, in an unzipping geometry, lower the Ttol to ~10 pN.[8] Fluorophore labeled TGTs anchored to a glass coverslip and imaged using high-resolution fluorescence microscopy have been used to study many pathways including integrin,[9] T cell receptor,[6b] and Notch receptor signaling.[7]
Molecular methods such as MTFM and TGT, hold great promise for force quantification and elucidation of molecular mechanism in mechanobiology, but they are performed on planar coverslips whereas physiological cell activities such as the immunological synapse (IS) formation and phagocytosis occur on non-planar geometries and have reported to depend on the curvature of the substrate.[10] According to recent work, this dependency may be attributed to enhanced mechanical activities such as actin polymerization at curved membranes.[11] To fill this gap in knowledge, we are aiming to develop a force sensor that allows one to investigate mechanotransduction on non-planar geometries. A number of groups, including Ning et al.,[12] Ingber et al.,[13] Theriot et al.[14] and Moraes et al.[15] have developed force-deformable microspheres to investigate cellular forces in processes that range from phagocytic engulfment to 3D cell spheroid mechanical homeostasis. However, these particles do not reveal the forces transmitted by individual receptor-ligand complexes, which is required to elucidate the molecular mechanisms of mechanobiology. A general problem that pertains to current molecular probes as well as these dispersible microparticle force sensors is their reliance on microscopy-based characterization, which is limited in throughput and at best, may allow for measuring tens to hundreds of samples per day. Ideally, molecular force measurements need to be increased in throughput to facilitate screening of biological samples and for efficient screening drugs that modulate mechanics.
Herein, to address these challenges, we developed a DNA-based microparticle tension sensor (μTS), where probes are immobilized on cell-sized dispersible particles that enables analysis of cell forces on non-planar interfaces as well as in high throughput by flow cytometry. We created μTSs displaying different ligands to engage cells that generate a fluorescence signal in response to forces transmitted through receptor-ligand bonds at the cell surface, which were then visualized by high resolution microscopy. Complementing force microscopy performed on planar substrates, μTS provides a novel geometric tool to measure cell forces generating on curved geometries, and thus may allow one to investigate mechanotransduction on curved interfaces and help understand how curvature influences mechanical events. We characterized TCR forces in a hybrid immunological synapse (IS) formed between μTSs and T-cells using confocal microscopy and found that force signals were concentrated in a ring- like pattern and colocalized with the enriched actin filaments. This finding is consistent with the centripetal force observed along the cell edge in micropillar array and the actin enrichment at the periphery of IS formed on support lipid bilayer.[16] In addition to 3D high resolution imaging, the cellular scale of μTS enables flow cytometry-based high throughput characterization. We applied this assay to measure platelet forces, revealing that integrin receptors transduce forces up to 56 pN. Since high throughput analysis is of significant interest for drug screening, we further applied μTS to investigate how drugs modulate cell traction forces. The mean fluorescence intensity of μTS was observed to decrease with increasing drug dose, showing the utility of μTS in screening drug that modulates cell mechanics. μTS measured mechanical inhibition (mechanical IC50) for Y-27632 and eptifibatide are 2.1 and 3.2 μM, respectively, for mouse platelets. These mechanoIC50 values are consistent with literature values determined using indirect readouts such as secondary messenger signaling and cell aggregation phenotypes.[17]
Results and Discussion
Design, synthesis and characterization of μTS.
μTSs were constructed by immobilizing molecular tension probes on cell-sized silica microparticles. Here, we chose fluorophore-quencher labeled TGTs (Figure S1) as tension probes because these provide an irreversible signal that accumulates with time, and thus generating greater levels of signal/noise compared to that of realtime MTFM probes. In principle, one can select a wide variety of microparticle materials that can be employed as a scaffold for μTS. The most desirable materials are monodisperse, show low autofluorescence, and afford facile and efficient chemical coupling to nucleic acids. Based on these criteria we used amine-modified silica beads with a diameter of 5 μm. We covalently linked the DNA probes to the particle using thiol-Michael reaction to immobilize thiolated-TGTs onto maleimide-modified spherical surfaces (Figure S2).[18] Recent measurements showed that this bond can withstand forces up to 100’s of pNs in aqueous environments.[19] By tuning the thiol position on the TGT, and hence controlling the geometry of anchoring (shearing versus unzipping), we tuned the Ttol of the 21 base pairs TGT from 12 pN in the unzipping geometry to 56 pN in the shearing geometry. Probe sensitivity was optimized by selecting fluorophore-quencher pairs with the highest quenching efficiency. Cy3BBHQ2 showed QE of ~95%, and thus generate a ~20-fold enhancement in signal upon mechanical denaturation (Figure S3). The TGT grafting chemistry was optimized such that the probe density on the spherical surfaces was 3660+/−200 molecules/μm2 (Figure S4). This probe density is markedly greater than the TCR receptor density on T cells, and is at the same order of magnitude as the density of αIIbβ3 integrin receptors on platelets, and thus guarantees effective ligand presentation and robust interaction between μTS and cells.[20]
Visualizing TCR mechanics in hybrid immunological synapse formed with spherical μTS.
Tension probes were first validated using lymphocytes cultured on planar surfaces. To study primary CD8+ T cell receptor forces, we employed the peptide major histocompatibility complex (pMHC) antigen that was loaded with the ovalbumin (OVA) derived 8-mer peptide: SIINFEKL (N4 peptide). The C-terminus of the pMHC antigen was modified with a biotin ligase sequence that allowed for conjugation to the TGT with biotin-streptavidin. In these experiments, naïve CD8+ cells were harvested from OVA-specific TCR transgenic mice (OT-1) and were then plated on glass coverslips presenting N4 peptide tagged 12 pN and 56 pN TGT probes. (Figure S5). Strong fluorescence (up to 5-fold over background) was exclusively observed underneath the cells engaged with 12 pN TGT surfaces, but not with the 56 pN TGT surfaces, indicating that TCR forces did not exceed 56 pN. Given that the thiolmaleimide anchored TGTs generated robust signal in response to TCR forces on planar surfaces, we next constructed μTS that displayed N4 peptide tagged TGT to investigate mechanical events occurring at the curved cell-μTS junction (Figure 1A). To facilitate sequential modifications and microscopy imaging, μTSs were immobilized on the glass coverslip through click reaction (Figure S6). Afterwards, naïve CD8+ T-cells were plated on μTS coated surface and incubated for 30 min to allow cells to form adhesions on μTS. By focusing on the z-position that roughly corresponds to the mid-section of the junction (~2.5 μm above the coverslip), strong turn-on signal (up to 6-fold over background) was observed at the junction between T-cell and 12 pN μTS (3 independent experiments, 20 μTS-cell binding events Figure 1B and 1C). Note that we did not detect significant tension signal for the 56 pN μTS particles, showing that even in the spherical geometry, TCR-pMHC forces do not achieve peak forces exceeding 56 pN. This experiment serves as an additional control since the thermal melting temperature of the 56 pN μTS is identical to that of the 12 pN μTS probes.
Figure 1.
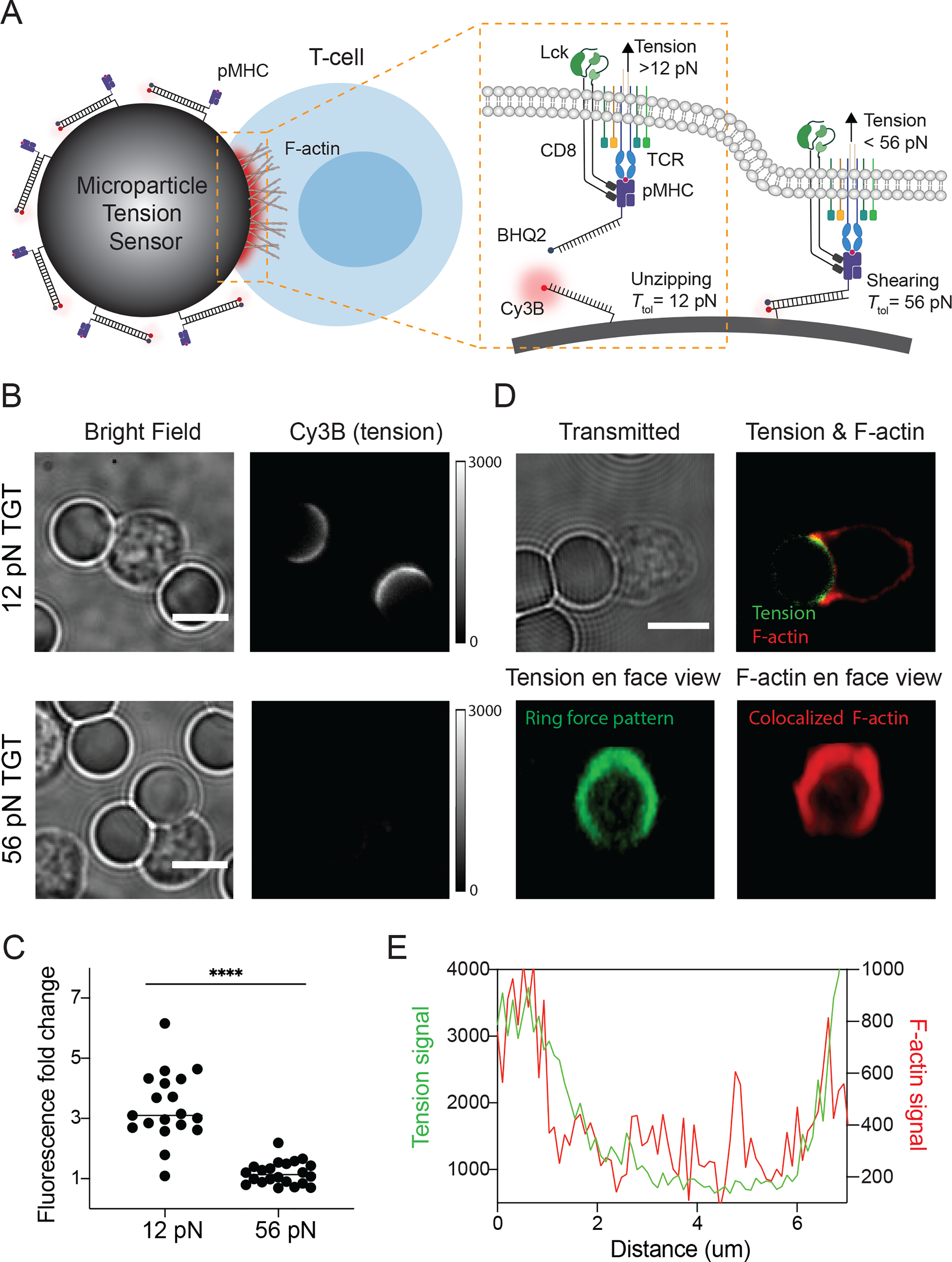
Mechano-imaging of TCR forces on spherical surfaces. (A) Schematic of μTS for mapping TCR forces transmitted through TCR-pMHC bonds at cell-μTS junctions. (B) Representative bright field and Cy3B fluorescence images showing adhesion and tension signals localized at junctions between T-cells and 12 pN μTS. (C) Plot showing fluorescence intensity fold change of junctions on μTS compared to background. Each dot represents single junctions from 3 different animals in independent experiments. **** indicates p<0.0001. (D) Representative confocal images showing ring-like tension signal (green) and F-actin signal (red) at T-cell-μTS synapse. (E) Linescans of F-actin signal (red line) and tension signal (green line) at the interface from junction shown in D. Scale bars = 5 μm.
The 12 pN tension distribution across the hybrid immunological synapse was further resolved by z-stack reconstruction of confocal images. As shown in Figure 1D, cells that encountered the μTS spread and engaged the particle surface that in some cases covered an area that was ~ 1/4 of the entire particle, as determined by the tension signal. Strikingly, the tension signal showed a ring-like spatial pattern that coincided with the cell perimeter, suggesting that TCR-pMHC mechanical events accumulate at the periphery of the T cell-antigen presenting cell junction. A similar geometric pattern was observed on planar surfaces modified with molecular tension sensors presenting pMHC ligands (Figure S3). This force pattern is consistent with our prior MTFM measurements as well as the centripetal forces observed along the cell edges on micropillar arrays.[6b, 16b, 21] Since actin polymerization and actomyosin contraction play important roles in force generation,[1b, 22] we next investigated the colocalization between actin filament and tension signal. After allowing cells to spread on the spherical and planar surfaces, cells were fixed and stained with the SiR-Actin stain (dye conjugated to F-actin stabilizing macrocycle). As expected, at the same z position, F-actin and tension signal were colocalized and both enriched at the edge of interface (Figure 1D–E and Figure S7). 3D view of F-actin obtained by combining z-stack images showed a ring-like pattern similar to the ring-shaped force signal pattern (Figure 1E and Movie S1).
Mapping platelet adhesion forces by μTS.
We next aimed to broaden the scope of the μTS platform by applying it to investigate force transmission through integrin receptors on a mouse platelet model. Platelets are small (1–2 micron) anuclear blood cells that play key roles in hemostasis including coagulation. Each platelet presents tens of thousands of integrin receptors (primarily αIIbβ3) that selectively bind to fibrinogen, the third most abundant protein in the blood plasma, which leads to platelet-to-platelet aggregation and contraction at the injury sites.[23] Our lab previously employed DNA hairpin-based MTFM and TGT probes to investigate integrin forces applied by human platelets, demonstrating that these forces range from 4.7 to 19.3 pN, with a subpopulation exceeding 19.3 pN when measured using the reversible MTFM probes.[24] The results from the TGT showed that platelet integrins can apply peak forces that exceed 56 pN. Wang et al. confirmed this conclusion, and by using turn-on TGTs, they showed that integrin receptors in canine platelets can shear and unzip the TGT probe, with greater signal in the unzipping mode.[25] However, these experiments were performed on isolated platelets (non-aggregated) engaged to planar glass surfaces. The collective aggregation of platelets that are bridged by fibrinogen will likely modulate the magnitude of molecular forces applied to ligands on a particle surface. Here we created μTS displaying cyclized RGD, which was conjugated to TGT top strand by Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) (Figure S2). The goal was to investigate aggregated mouse platelet integrin mechanics on a non-planar geometry (Figure 2A).
Figure 2.
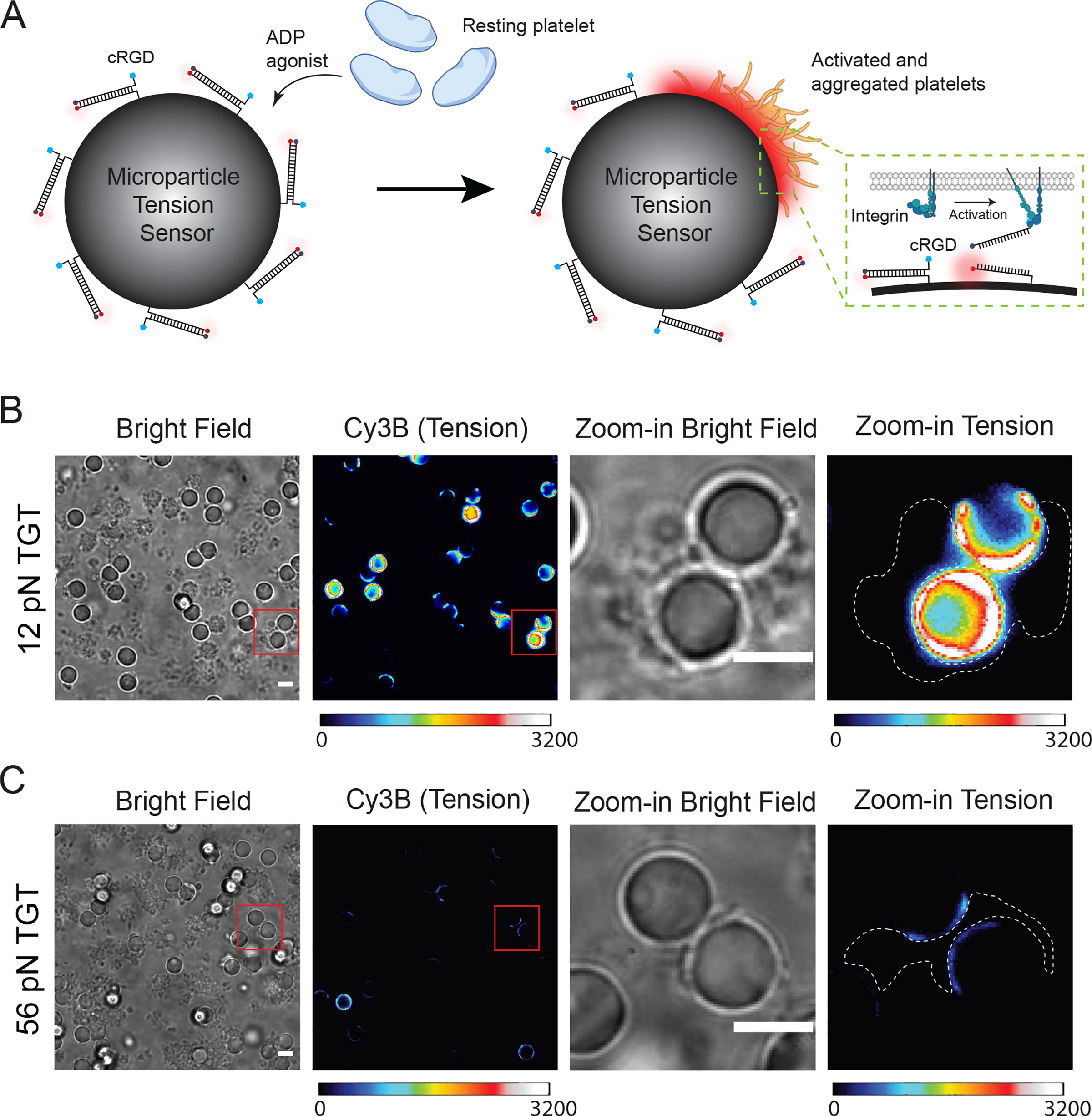
Visualizing integrin forces with μTS. (A) Schematic showing how μTS reveals platelet integrin tension through cRGD coated DNA probes on the μTS. (B) Representative bright field and tension images of 12 pN μTS-platelet complexes. The platelets were mixed with μTS for 30 min and then imaged. (C) Representative bright field and tension images of 56 pN μTS-platelet complexes. The images in B and C are displayed at identical contrasts and can be compared directly. Scale bars = 5 μm.
In a typical experiment, μTS were incubated with millions of mouse platelets at approximately 1:50 ratio in Tyrode buffer supplemented with 10 μM adenosine diphosphate (ADP), 2 mM MgCl2 and 1 mM CaCl2 for 30 min to allow platelets to adhere and form aggregates on μTS surface (Figure 2B–C). Afterwards, forces transmitted by the platelet integrin receptors to μTS were visualized using epi fluorescence microscopy. As shown in Figure 2B, up to a 10-fold fluorescence increase was observed with the 12 pN μTS. The tension signal was primarily localized to the particle-platelet junctions. In contrast, when we employed the shearing, 56 pN μTS, there was weak tension signal suggesting that only small subset of receptors transmitted peak forces F > 56 pN in these conditions. The brightest particles in the 56 pN case showed a 3-fold intensity increase (Figure 2C). These results indicate that platelets within aggregate transmit similar integrin traction forces compared to the levels observed for single platelets engaged to planar glass slides. In addition, this result dispels the prediction that untether beads engaged to aggregates of platelets linked by fibrinogen would experience diminished tension.
Quantifying integrin forces in high throughput by μTS.
Since μTSs efficiently interact with platelets and exhibit strong fluorescence signal in response to integrin forces, we next wondered whether the force signal recorded on the surface of individual particles could be quantified in a high throughput fashion using flow cytometry. In principle, the micrometer diameter of the probes strongly suggests that it would be amenable to flow cytometry analysis. Prior work by Tom Soh and colleagues showed that flow cytometry of microparticles can be used for screening libraries of nucleic acids to identify improved aptamers and deoxyribozymes.[26] Moreover, flow-based analysis is widely used in immunology and cell biology to characterize heterogenous cell populations.[27] This precedent provides the rationale and motivation for pursuing flow-based analysis to study molecular mechanobiology. To the best of our knowledge, this approach would represent the first example of using flow cytometry to quantify molecular mechanics of cells in high throughput.
In the suspension, the number of platelets was significantly larger than that of μTS, and the aggregate size was heterogeneous, gating discrete stoichiometries of cell-platelet assemblies was not possible. Therefore, after 30 min incubation at room temperature, we lysed all platelet aggregates and then collected μTS particles for flow cytometry analysis. After adding lysis buffer, all aggregates disappeared rapidly (~min) (Figure 3A and Figure S8). μTS fluorescence remained stable in the lysis buffer for at least 15 min, indicating that this buffer did not lead to DNA denaturation (Figure S8). In light of this, we next acquired 10,000 μTS events through flow cytometry and quantified the force signal by comparing the mean fluorescence intensity (MFI) of these events to that of μTS that were not incubated with platelets but subjected to the lysis buffer protocol (Figure 3B). We found that the MFI of 12 pN μTS monomers increased 90±7% after binding to platelets while 56 pN μTS monomers only increased 39±9% (3 independent experiments, Figure 3C). This result is highly consistent with microscopy readouts (Figure S9). As a negative control, μTS coated with TGT tension probe presenting cyclized RAD peptide weakly engaged platelets and exhibited negligible fluorescence change (Figure S10). These results show that platelet integrin receptors apply peak forces in the range of 12 to 56 pN, with a small subset of receptors transmitting forces that exceed 56 pN. To further demonstrate the advantage of μTS flow readout compared to that of microscopy-based methods, we also tested the potential for barcoding and multiplexing multiple levels of force in a single experiment (Figure S11). Here, we spectrally encoded the 12 pN and 56 pN μTS probes and mixed these with platelets. The barcoded μTS confirmed our single color experiments with 12 pN signal far exceeding that of the 56 pN signal. Past AFM work showed that the contractile forces applied by platelets are proportional to the stiffness of the substrate,[28] and given that molecular probes on the μTS experience similar forces to probes anchored on planar glass slides, this suggests that the platelet integrins are sensing the local mechanical properties of the silica particle and forces are not dampened due to having the particles in suspension.
Figure 3.
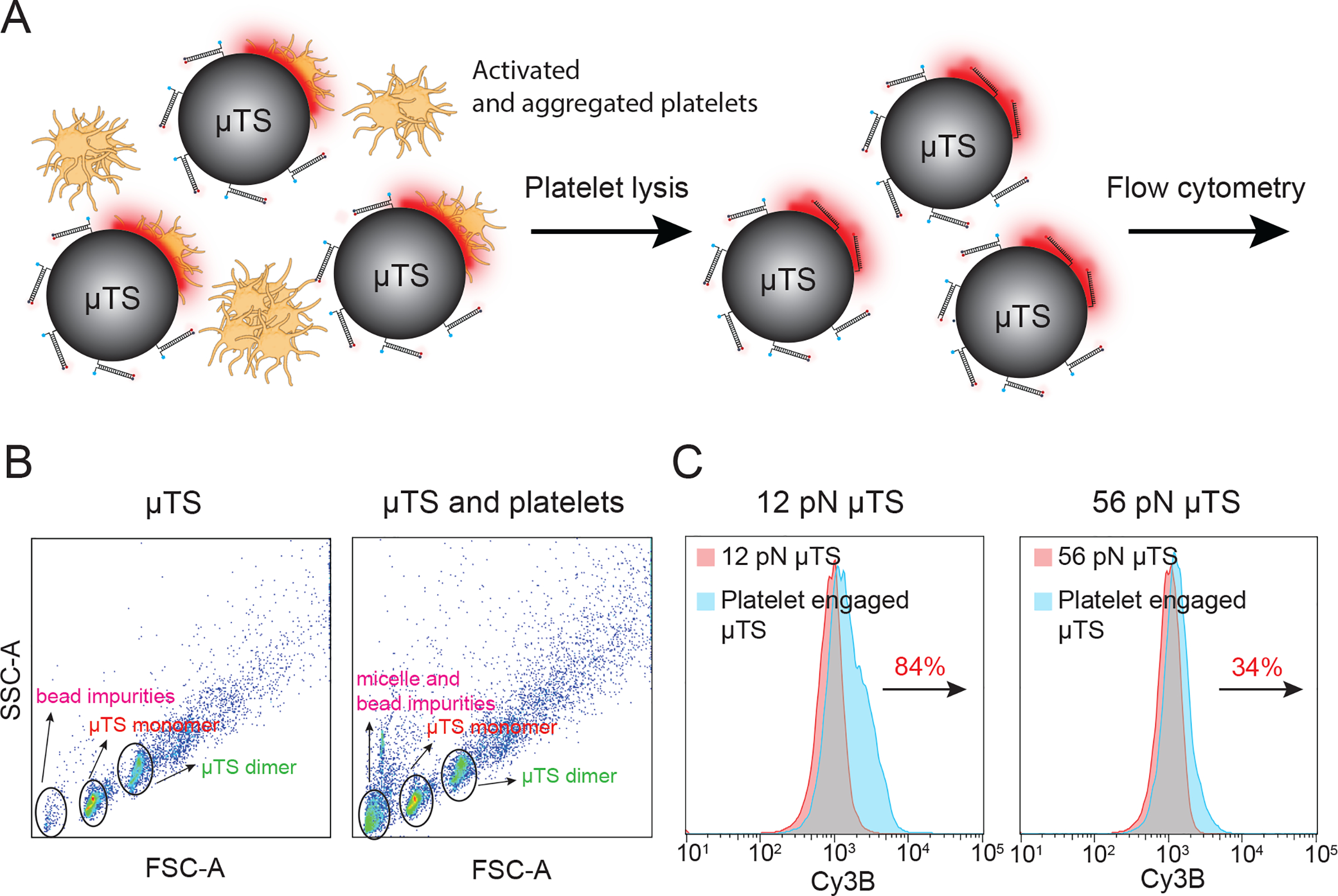
Flow cytometry-based characterization of integrin forces. (A) Schematic showing the procedure used for preparing platelet samples for μTS flow cytometry analysis. (B) SSC vs FSC plots of μTS control sample (left) and platelet-engaged μTS (right). Based on a series of controls, events were assigned as μTS monomers (red), μTS dimers (green), and impurities (defective μTS particles or other debris shown in pink). (C) Fluorescence histograms of gated events of μTS monomers for the 12 and 56 pN probes. The red population corresponds to the control μTS not incubated with platelets while the blue indicates μTS that engaged platelets. The percent change indicates the change in the mean fluorescence intensity of μTS. N=10000 events for each group.
Mechanopharmacology using the μTS platform: Testing how anti-platelet drugs impact platelet integrin forces.
Since probes amenable to high throughput analysis are of significant interest for their potential application in drug screening, we next aimed to demonstrate the suitability of μTS in measuring how platelet contractile forces can be modulated using drugs. It is notable that platelet traction forces have been shown as a prognostic marker of coagulation both in trauma patients as well as for hereditary mutations in coagulation cascades,[29] and hence μTS readout offers physiological relevance to clotting. Here we tested two anti-platelet drugs that inhibit platelet mechanics via different mechanisms. Y-27632 inhibits Rho-associated protein kinase (ROCK) and diminishes actomyosin contractility while the second drug eptifibatide is FDA approved to treat acute coronary syndrome, and binds competitively to aIIbb3 integrin to block the interaction between integrins and adhesive ligands. To investigate the effects of these drugs on tension signals, platelets were pretreated with different concentrations of drugs for 20 min and then cultured with 12 pN μTS for 1 hour, which was followed by microscopy and flow cytometry characterization. After Y27263 drug treatment, fluorescence signals at the μTS-platelet junctions decreased with increasing drug concentration (Figure 4A). Consistently, flow cytometry data revealed that the MFI of μTS displayed a dose dependent relationship with a relative IC50=2.08 μM (Figure 4B). However, platelet-to-platelet aggregation was not affected by Y27632 (Figure S12), indicating that ROCK is an important but dispensable factor in platelet aggregation. The aggregation was maintained by the highly concentrated ADP in the suspension, which bound to ADP receptors such as P2Y12 on the platelet membrane to trigger platelet aggregation.[30] Additionally, past work showed that ADP could induce a burst of talin activation to trigger inside-out integrin activation, which leads to platelet activation and aggregation.[31] The tension signal of eptifibatide treated platelets exhibited similar dose-dependent relationship with IC50=3.24 μM (Figure 4C). However, in contrast to Y27632 which can only abolish the tension signal up to 60%, eptifibatide is able to inhibit 80 % of the tension signal, indicating that eptifibatide is a more potent drug compared with Y27632 in modulating applied integrin forces. Note, in addition to force signal changes, aggregation and the adhesion of eptifibatide treated platelets was significantly reduced despite the presence of ADP agonist (Figure 4A and Figure S10). This is because the blockade of integrin directly inhibits the interaction with μTS and surrounding platelets, confirming the necessity of ligand binding in integrin activation and platelet aggregation.
Figure 4.
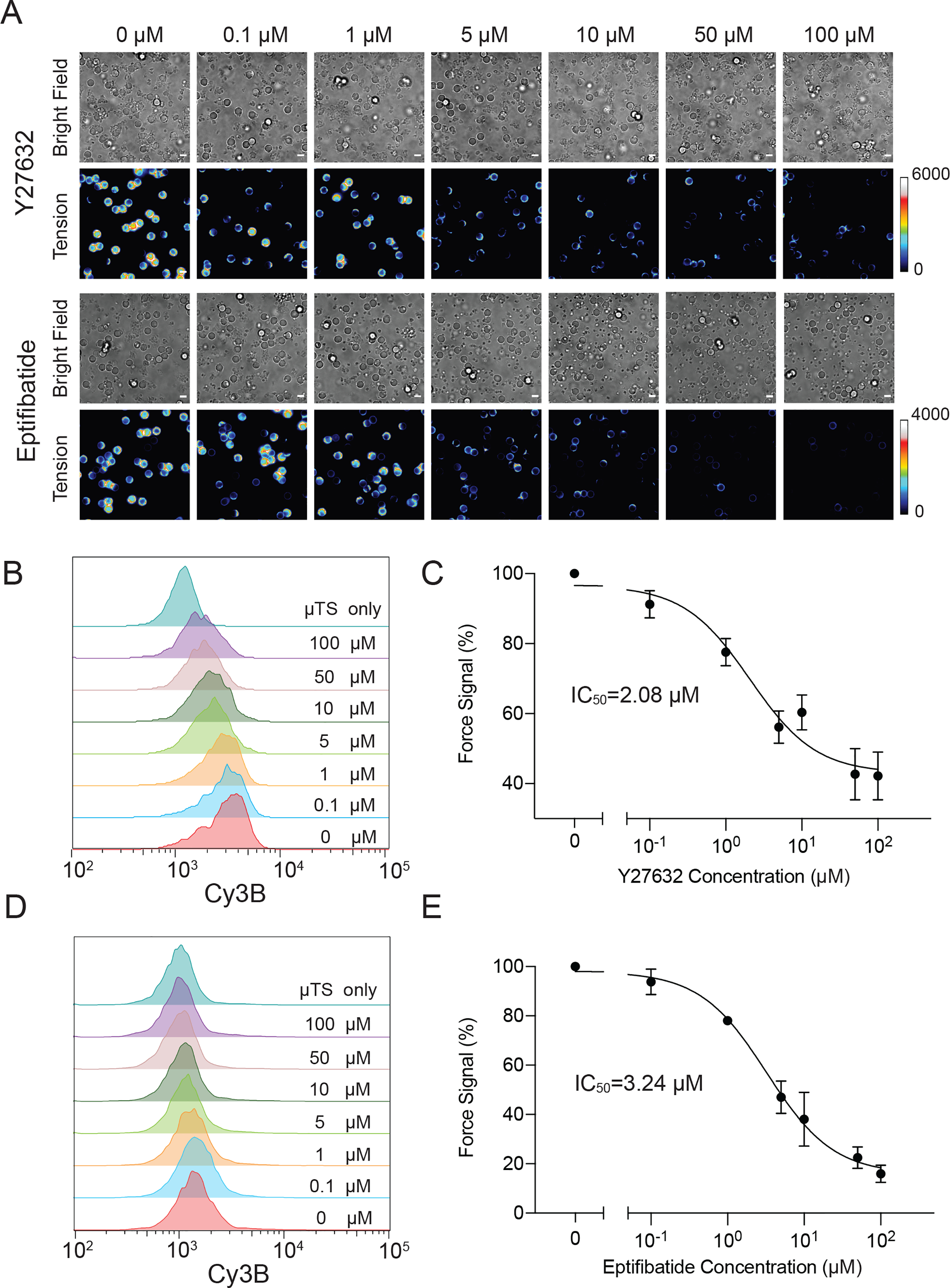
Proof-of-concept demonstration of measuring dose-response curve for drugs that modulate platelet mechanics. (A) Representative bright field and fluorescence images showing a mixture of platelets with μTS probes that were allowed to interact for 1 hr and then seeded on a substrate. Platelets were pretreated with a range of concentrations of Y27632 and eptifibatide (0–100μM) for 20 min. (B) Flow-cytometry based histograms of fluorescence intensity of μTS that engaged platelets treated with different concentrations of Y27632. (C) Dose-response curve plotting the tension signal obtained from flow cytometry plotted against the log concentration of Y27632. (D) Flow-cytometry based histograms of fluorescence intensity of μTS probes that engaged platelets pretreated with different concentrations of eptifibatide. (E) Dose-response curve plotting the tension signal obtained from flow cytometry against the log concentration of eptifibatide. Each histogram is plotting N = 10000 events. Error bars show the standard error of the mean from three independent measurements from three animals. Scale bars = 5 μm
Conclusion
DNA-based microparticle tension sensors (μTS) show great potential in investigating cell receptor mechanics. In contrast to planar coverslip-based force measurement assays, for T cells, the μTS features a spherical surface that allows the characterization of molecular forces at this curved interface. We applied μTS to investigate T-cell and platelet forces, revealing that TCR and integrin receptors transduce forces ranging from 12 to 56 pN, with a subpopulation of integrin receptors experiencing F>56 pN. Impressively, we observed a highly resolved 3D ring-like force pattern at the hybrid synapse formed between μTS and T-cell, showing that T-cell receptors are mechanically active at the periphery of cells adhered on non-planar surfaces. To the best of our knowledge, no study to date has mapped molecular forces on non-planar geometries. Note that microparticles have been instrumental in investigating actin distribution,[32] receptor-ligand interactions,[33] and traction forces[14] on non-planar geometries, and thus μTS may complement previous methods and allow one to investigate these mechanical events with molecular detail. In principle, the curvature of μTS can be tuned by varying the size of the microparticle template, suggesting that this approach has the potential to facilitate the study of how cell curvature can modulate cell activities. In addition to the non-planar geometry, another advantage of μTS is its micrometer diameter, which enables flow cytometry-based high throughput characterization. Through flow cytometry, hundreds of thousands of force signalassociated events were acquired and analyzed within minutes, which is in contrast to microscope-based arrays that are orders of magnitude slower in throughput and would take ~ 1 day for analyzing hundreds of events even upon integration of automated robotic imaging. In addition, flow cytometers with up to 14 fluorescence channels would allow for simultaneous and multiplexed force investigation reporting the magnitude of tension transmitted by different receptor-ligand pairs on the cell surface. Complementing this future direction, recently developed imaging flow cytometry instruments generate high resolution images of each particle, thus providing spatial information in a massively high throughput manner in comparison to conventional confocal or TIRF microscopy that are used in high-content screening assays.[34] The potential application of the μTS platform in drug screening was demonstrated by investigating the effect of two anti-platelet drugs. The IC50 values for Y27632 and eptifibatide were 2.08 and 3.24 μM, respectively, which are consistent with literature reported values of ~1 μM for both of these drugs.[17a, 35] Note that the literature values were recorded using secondary messenger assays (aggregation, morphology or calcium influx) and thus our molecular probes offer a more precise measure of platelet contractility. The μTS platform still has important limitations. First, the μTS core is comprised of silica which is a hard material compared to that of cells and tissue. Also, the S/N depends on the stoichiometry between cells and beads, as the fraction of the μTS surface that engages cells tunes the strength of the signal. We anticipate that future work using amplification strategies and gel-based particles may address these limitations. In summary, we demonstrated that μTS is an efficient platform for molecular force quantification capable of visualizing molecular force on non-planar geometry in high spatial resolution as well as determining its magnitude in high throughput.
Supplementary Material
Acknowledgements
This work was supported by NIH R01 GM124472 (K.S.), NIH R01 GM131099 (K.S.) and NIH HL082808 (R.L.). V.P.-Y.M. was supported by NCI K00 CA223074. We acknowledge the support from Emory Flow Cytometry Core. We thank the NIH Tetramer Facility for pMHC ligands. Some illustrations are adapted from Biorender.com templates. This study was supported, in part, by the Emory Comprehensive Glycomics Core.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Publisher's Disclaimer: This manuscript has been accepted after peer review and appears as an Accepted Article online prior to editing, proofing, and formal publication of the final Version of Record (VoR). This work is currently citable by using the Digital Object Identifier (DOI) given below. The VoR will be published online in Early View as soon as possible and may be different to this Accepted Article as a result of editing. Readers should obtain the VoR from the journal website shown below when it is published to ensure accuracy of information. The authors are responsible for the content of this Accepted Article.
References
- [1].a Fletcher DA, Mullins RD, Nature 2010, 463, 485–492; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Murrell M, Oakes PW, Lenz M, Gardel ML, Nat Rev Mol Cell Biol 2015, 16, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a Roca-Cusachs P, Conte V, Trepat X, Nat Cell Biol 2017, 19, 742–751; [DOI] [PubMed] [Google Scholar]; b Polacheck WJ, Chen CS, Nat Methods 2016, 13, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K, J Cell Biol 1994, 127, 1957–1964; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Koch TM, Munster S, Bonakdar N, Butler JP, Fabry B, PLoS One 2012, 7, e33476; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Legant WR, Miller JS, Blakely BI, Cohen DM, Genin GM, Chen CS, Nature Methods 2010, 7, 969–971; [DOI] [PMC free article] [PubMed] [Google Scholar]; d Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS, Proc Natl Acad Sci U S A 2013, 110, 881–886; [DOI] [PMC free article] [PubMed] [Google Scholar]; e Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS, Proc Natl Acad Sci U S A 2003, 100, 1484–1489; [DOI] [PMC free article] [PubMed] [Google Scholar]; f Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan-Jones A, Margadant F, Mege RM, Lim CT, Voituriez R, Ladoux B, Nat Commun 2015, 6, 7525; [DOI] [PMC free article] [PubMed] [Google Scholar]; g Schwarz US, Soine JR, Biochim Biophys Acta 2015, 1853, 3095–3104. [DOI] [PubMed] [Google Scholar]
- [4].a Stabley DR, Jurchenko C, Marshall SS, Salaita KS, Nat Methods 2011, 9, 64–67; [DOI] [PubMed] [Google Scholar]; b Liu Y, Yehl K, Narui Y, Salaita K, J Am Chem Soc 2013, 135, 5320–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Galior K, Liu Y, Yehl K, Vivek S, Salaita K, Nano Lett 2016, 16, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a Zhang Y, Ge C, Zhu C, Salaita K, Nat Commun 2014, 5, 5167; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K, Proc Natl Acad Sci U S A 2016, 113, 5610–5615; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ma R, Kellner AV, Ma VP, Su H, Deal BR, Brockman JM, Salaita K, Proc Natl Acad Sci U S A 2019, 116, 16949–16954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang X, Ha T, Science 2013, 340, 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mosayebi M, Louis AA, Doye JPK, Ouldridge TE, ACS Nano 2015, 9, 11993–12003. [DOI] [PubMed] [Google Scholar]
- [9].a Zhao Y, Pal K, Tu Y, Wang X, J Am Chem Soc 2020, 142, 6930–6934; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang X, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang Y, Ha T, Biophys J 2015, 109, 2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a Champion JA, Mitragotri S, Proc Natl Acad Sci U S A 2006, 103, 4930–4934; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Richards DM, Endres RG, Proc Natl Acad Sci U S A 2016, 113, 6113–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lou HY, Zhao W, Li X, Duan L, Powers A, Akamatsu M, Santoro F, McGuire AF, Cui Y, Drubin DG, Cui B, Proc Natl Acad Sci U S A 2019, 116, 23143–23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mohagheghian E, Luo J, Chen J, Chaudhary G, Chen J, Sun J, Ewoldt RH, Wang N, Nat Commun 2018, 9, 1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Campàs O, Mammoto T, Hasso S, Sperling RA, O’Connell D, Bischof AG, Maas R, Weitz DA, Mahadevan L, Ingber DE, Nat Methods 2014, 11, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vorselen D, Wang Y, de Jesus MM, Shah PK, Footer MJ, Huse M, Cai W, Theriot JA, Nat Commun 2020, 11, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee W, Kalashnikov N, Mok S, Halaoui R, Kuzmin E, Putnam AJ, Takayama S, Park M, McCaffrey L, Zhao R, Leask RL, Moraes C, Nat Commun 2019, 10, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD, Proc Natl Acad Sci U S A 2007, 104, 20296–20301; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jin W, Tamzalit F, Chaudhuri PK, Black CT, Huse M, Kam LC, Proc Natl Acad Sci U S A 2019, 116, 19835–19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a Ishizaki T, Uehata M, Tamechika I, Keel J, Nonomura K, Maekawa M, Narumiya S, Mol Pharmacol 2000, 57, 976–983; [PubMed] [Google Scholar]; b Paul BZS, Daniel JL, Kunapuli SP, J Biol Chem 1999, 274, 28293–28300. [DOI] [PubMed] [Google Scholar]
- [18].Zimmermann JL, Nicolaus T, Neuert G, Blank K, Nat Protoc 2010, 5, 975–985. [DOI] [PubMed] [Google Scholar]
- [19].Huang W, Wu X, Gao X, Yu Y, Lei H, Zhu Z, Shi Y, Chen Y, Qin M, Wang W, Cao Y, Nat Chem 2019, 11, 310–319. [DOI] [PubMed] [Google Scholar]
- [20].a Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C, Nature 2010, 464, 932–936; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wagner CL, Mascelli MA, Neblock DS, Weisman HF, Coller BS, Jordan RE, Blood 1996, 88, 907–914. [PubMed] [Google Scholar]
- [21].Ma VP, Liu Y, Blanchfield L, Su H, Evavold BD, Salaita K, Nano Lett 2016, 16, 4552–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a Ma VP, Liu Y, Yehl K, Galior K, Zhang Y, Salaita K, Angew Chem Int Ed Engl 2016, 55, 5488–5492; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Footer MJ, Kerssemakers JW, Theriot JA, Dogterom M, Proc Natl Acad Sci U S A 2007, 104, 2181–2186; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Huse M, Nat Rev Immunol 2017, 17, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Qiu Y, Brown AC, Myers DR, Sakurai Y, Mannino RG, Tran R, Ahn B, Hardy ET, Kee MF, Kumar S, Bao G, Barker TH, Lam WA, Proc Natl Acad Sci U S A 2014, 111, 14430–14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang Y, Qiu Y, Blanchard AT, Chang Y, Brockman JM, Ma VP, Lam WA, Salaita K, Proc Natl Acad Sci U S A 2018, 115, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang Y, LeVine DN, Gannon M, Zhao Y, Sarkar A, Hoch B, Wang X, Biosens Bioelectron 2018, 100, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang J, Yu J, Yang Q, McDermott J, Scott A, Vukovich M, Lagrois R, Gong Q, Greenleaf W, Eisenstein M, Ferguson BS, Soh HT, Angew Chem Int Ed Engl 2017, 56, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].a Ahmed R, Omidian Z, Giwa A, Cornwell B, Majety N, Bell DR, Lee S, Zhang H, Michels A, Desiderio S, Sadegh-Nasseri S, Rabb H, Gritsch S, Suva ML, Cahan P, Zhou R, Jie C, Donner T, Hamad ARA, Cell 2019, 177, 1583–1599 e1516; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, Topfner N, Keyser UF, Mansfeld J, Fischer-Friedrich E, Guck J, Nat Methods 2015, 12, 199–202, 194 p following 202. [DOI] [PubMed] [Google Scholar]
- [28].Lam WA, Chaudhuri O, Crow A, Webster KD, Li TD, Kita A, Huang J, Fletcher DA, Nat Mater 2011, 10, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a Ting LH, Feghhi S, Taparia N, Smith AO, Karchin A, Lim E, John AS, Wang X, Rue T, White NJ, Sniadecki NJ, Nat Commun 2019, 10, 1204; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Myers DR, Qiu Y, Fay ME, Tennenbaum M, Chester D, Cuadrado J, Sakurai Y, Baek J, Tran R, Ciciliano JC, Ahn B, Mannino RG, Bunting ST, Bennett C, Briones M, Fernandez-Nieves A, Smith ML, Brown AC, Sulchek T, Lam WA, Nat Mater 2017, 16, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bandyopadhyay SK, Azharuddin M, Dasgupta AK, Ganguli B, SenRoy S, Patra HK, Deb S, Front Bioeng Biotechnol 2019, 7, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sun Z, Costell M, Fassler R, Nat Cell Biol 2019, 21, 25–31. [DOI] [PubMed] [Google Scholar]
- [32].Barger SR, Reilly NS, Shutova MS, Li Q, Maiuri P, Heddleston JM, Mooseker MS, Flavell RA, Svitkina T, Oakes PW, Krendel M, Gauthier NC, Nat Commun 2019, 10, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bakalar MH, Joffe AM, Schmid EM, Son S, Podolski M, Fletcher DA, Cell 2018, 174, 131–142 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Weiss LE, Shalev Ezra Y, Goldberg S, Ferdman B, Adir O, Schroeder A, Alalouf O, Shechtman Y, Nat Nanotechnol 2020, 15, 500–506. [DOI] [PubMed] [Google Scholar]
- [35].Wang X, Dorsam RT, Lauver A, Wang H, Barbera FA, Gibbs S, Varon D, Savion N, Friedman SM, Feuerstein GZ, J Pharmacol Exp Ther 2002, 303, 1114–1120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


