Figure 3. Pooled shRNA library screening indicates glycolysis inhibition sensitizes iKras cells to MEK inhibition.
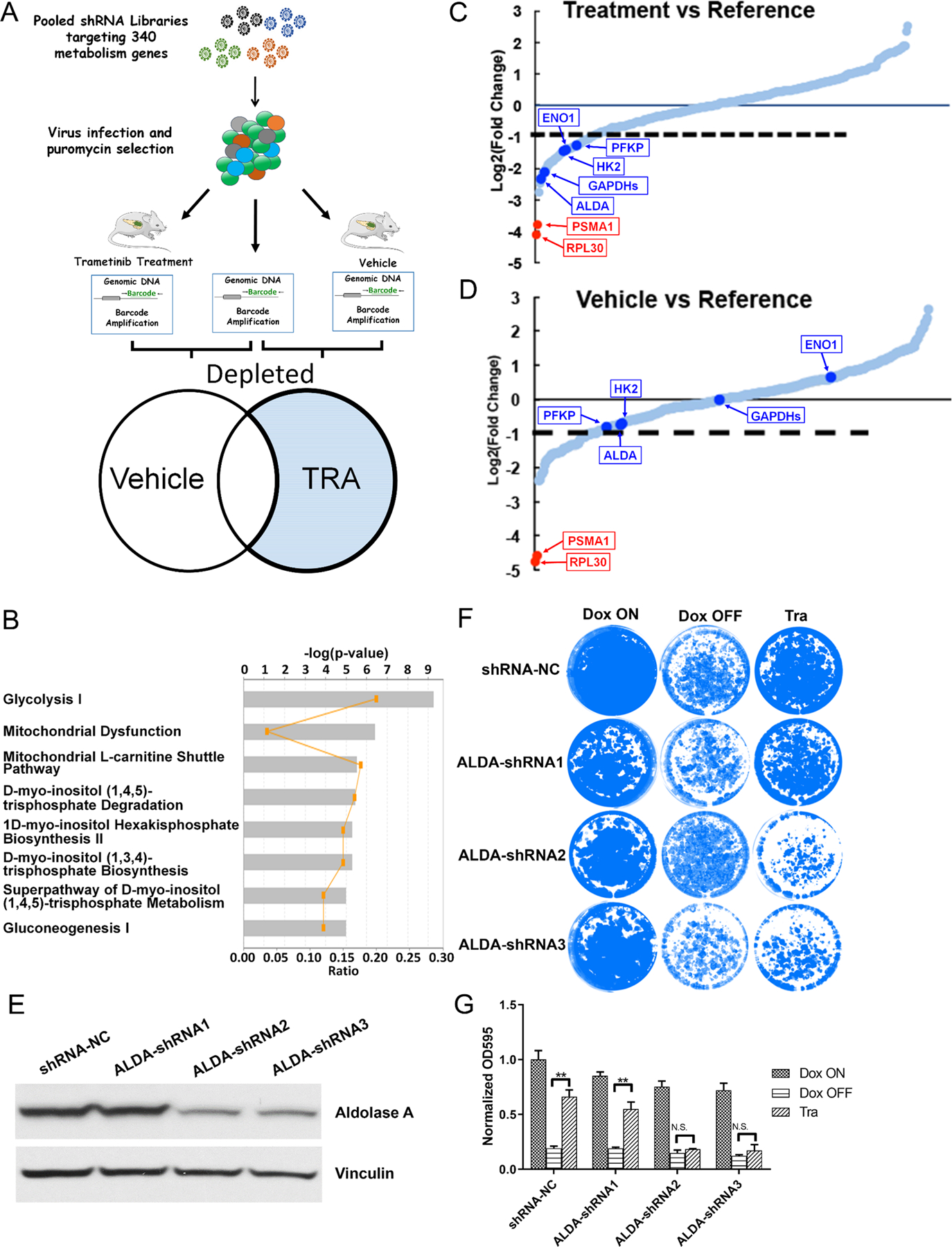
(A) Schematic illustration of pooled shRNA library screening in vivo. (B) IPA analysis of the depleted genes in shRNA library screen. (C-D) Relative abundance of each gene in the library in vehicle- or trametinib-treated tumors. Red dots: two positive control Rpl30 and Psma1; Blue Dots: five genes involved in glycolysis flux. (E) Knockdown of AldoA in iKras cell by shRNA. (F) Crystal violet staining of AldoA knockdown cell upon treatment of Dox ON, Dox OFF or 25 nM Tra for 4 days. (G) Quantification of crystal violet staining in F (n=3, Mean ± SD).
