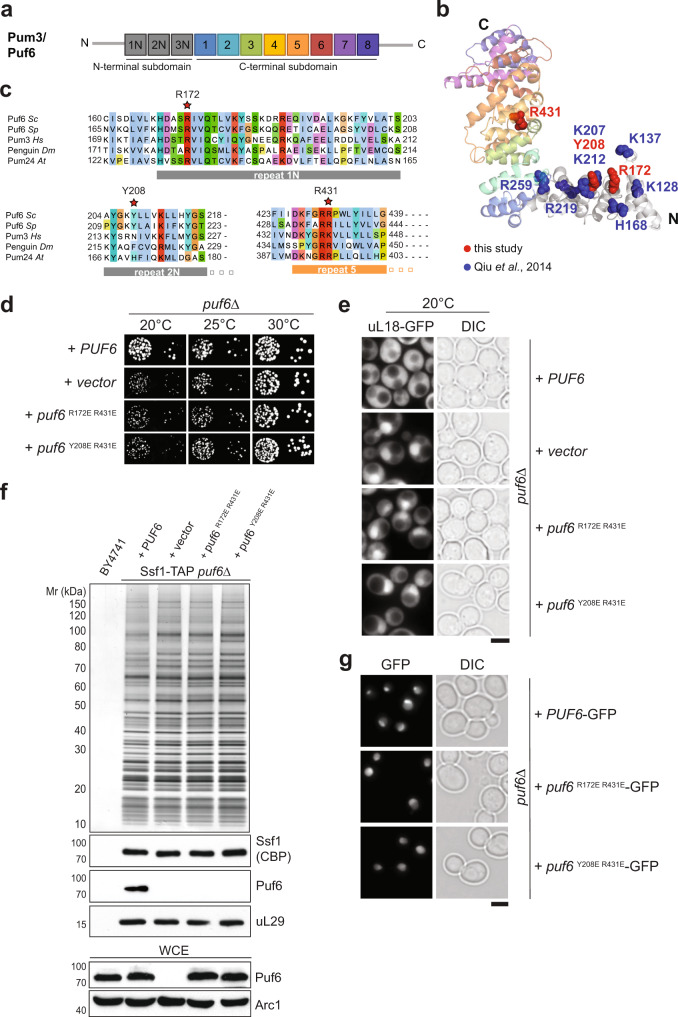
Fig. 4. The Pumilio repeat domain recruits Puf6 to 60S pre-ribosomes.
a Repeat organization of an atypical Pumilio repeat domain-containing Pum3/Puf6. b Cartoon representation of Puf6 homology model, visualized by PyMoL. The structure modeling of Puf6 was performed by using PHYRE2 server77. Of the 656 residues of Puf6, 118–654 residues could be modeled with 100% confidence using Pum3/Puf-A crystal structure (PDB ID: 4WZW) as the preferred template. Substituted residues within this study are shown in red. Previously substituted residues with altered affinities to nucleic acid substrates are shown in blue35. c Partial sequence alignment of Puf6—non-canonical PUF-repeat 1, 2, and canonical repeat 5 from Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Homo sapiens (Hs), Drosophila melanogaster (Dm) and Arabidopsis thaliana (At). The residues indicated by a red asterisk were substituted in yeast Puf6 for functional studies: R172, Y208, and R431 corresponding to R181, N217, and K440 in human Pum3/Puf-A35 d puf6∆ cells expressing WT-Puf6, and the indicated puf6 mutants were spotted in serial 10-fold dilutions on selective minimal medium plates and grown at indicated temperatures for 3–5 days. e The indicated strains expressing the 60S reporter, uL18-GFP, were grown at 20 °C till mid-log phase. Localization of uL18-GFP was analysed by fluorescence microscopy. Scale bar = 5 µm. f Ssf1-TAP was isolated from PUF6, puf6Δ and the indicated puf6 mutants. The TAP eluates were separated on a NuPAGE 4–12% Bis-Tris gradient gel and analyzed by Silver staining and Western blotting using antibodies directed against the bait (Ssf1-CBP) and Puf6. The r-protein uL29 (yeast Rpl35) was used as a loading control. Prior to TAP, whole-cell extracts (WCE) from the indicated yeast strains were separated by SDS-PAGE and analyzed by Western blotting using a Puf6-specific antibody. Arc1 was used as a loading control. g Yeast cells expressing C-terminally GFP tagged variants of Puf6 and puf6 mutants were grown to mid-log phase and analysed by fluorescence microscopy. Scale bar = 5 µm. Source data are provided as a Source Data file.