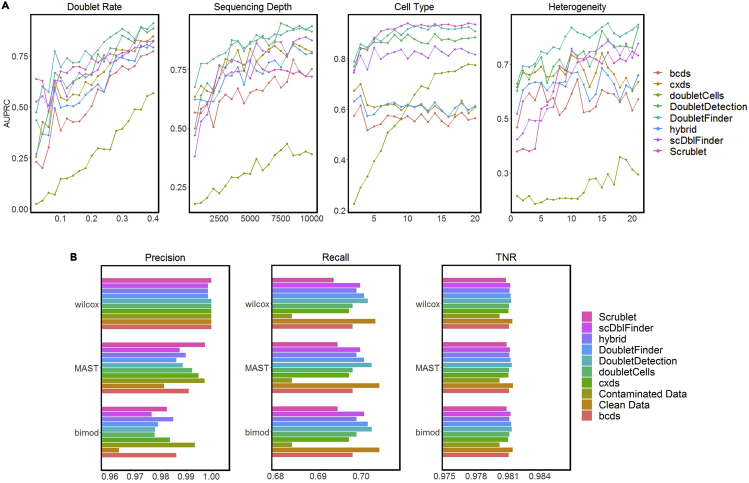
Figure 4.
Evaluation of doublet-detection methods using four simulation studies, and the effects of doublet detection on DE gene analysis
(A) AUPRC of each method in four simulation settings: varying doublet rates (from 2% to 40% with a step size of 2%), varying sequencing depths (from 500 to 10,000 UMI counts per cell, with a step size of 500 counts), varying numbers of cell types (from 2 to 20 with a step size of 1), and 20 heterogeneity levels, which specify the extent to which genes are differentiated between two cell types.
(B) Precision, recall, and TNR by each of three DE methods: Wilcoxon rank-sum test (wilcox), MAST, and likelihood-ratio test (bimod) after each doublet-detection method is applied to a simulated dataset; for negative and positive controls, we included the DE accuracies on the contaminated data with 40% doublets and the clean data without doublets.