Figure 10.
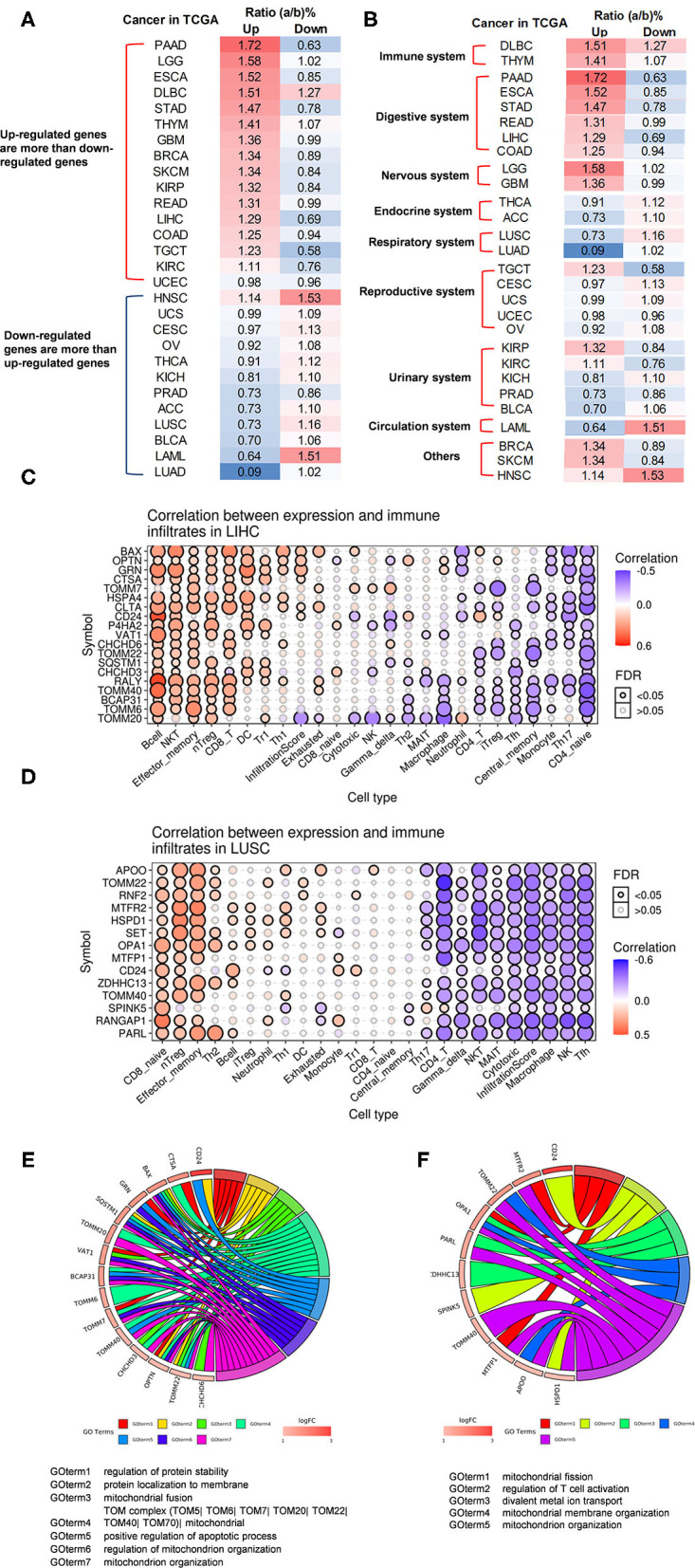
The ratio of upregulated and downregulated OCRGs has different dominance in tumors of different systems, and differentially expressed OCRGs are correlated with immune infiltrates in liver hepatocellular carcinoma (LIHC) and lung squamous cell carcinoma (LUSC). (A) Tumors are listed according to the ratio of upregulated and downregulated OCRGs. The results show that in pancreatic adenocarcinoma (PAAD), brain lower-grade glioma (LCC), and esophageal carcinoma (ESCA), these 15 cancers [from PAAD to uterine corpus endometrial carcinoma (UCEC)] upregulated OCRGs more than the downregulated OCRGs; in lung adenocarcinoma (LUAD), adrenocortical carcinoma (ACC), and acute myeloid leukemia (LAML), these 13 cancers (from HNSC to LUAD) downregulated OCRGs more than the upregulated genes. In order to rule out the influence of the overall expression profile on our interest OCRGs, we take the ratio of number of OCRGs and number of total differently expressed genes as the comparison criteria for the number of upregulated and downregulated genes. Result show that 16 cancers have upregulated OCRG percentage >1% and that 12 cancers have upregulated OCRGs percentages <1%. (B) Expression of OCRGs in tumors was roughly classified according to the system. The numbers of upregulated OCRGs are more than downregulated genes in tumors of immune system, digestive system, and nervous system. The numbers of downregulated OCRGs are more than upregulated OCRGs in tumors of the endocrine system, respiratory system, and circulation system and most tumors of the reproductive system and urinary system. In breast invasive carcinoma (BRCA) and skin cutaneous melanoma (SKCM), the numbers of upregulated OCRGs are more than those of downregulated OCRGs. In head and neck squamous cell carcinoma (HNSC), the numbers of upregulated OCRGs are less than those of downregulated OCRGs. (C) Almost 19 upregulated OCRGs in LIHC are positively correlated with B cell, NKT, effector memory, nTreg, CD8 T, DC, Tr1, and Th1 infiltration but negatively correlated with macrophage, neutrophil, CD4 T, iTreg, central memory, monocyte, Th17, and CD4 naïve cell infiltration. Expressions of BAX, OPTN, GRN, CTSA, CLTA, and CD24 are positively correlated with infiltration score, and expression of TOMM20 is negatively correlated with infiltration score. (D) Fourteen upregulated OCRGs in LUSC are positively correlated with CD8 T, nTreg, and effector-memory cell infiltration and negatively correlated with CD4 T, gamma-delta, NKT, MAIT, cytotoxic macrophage, NK, and Tfh cell infiltration. Expressions of all 14 OCRGs are negatively correlated with infiltration score. Correlations between expression of upregulated OCRGs and immune infiltrates were analyzed in LIHC and LUSC in Gene Set Cancer Analysis (GSCA) database (http://bioinfo.life.hust.edu.cn/GSCA/#/immune). (E) Fifteen out 19 upregulated OCRGs in LIHC enriched in seven significant pathways including regulation of protein stability, protein localization to membrane, mitochondrial fusion, TOM mitochondrial, positive regulation of apoptotic process, regulation of mitochondrion organization, and mitochondrion organization. More OCRGs enriched in TOM complex mitochondrial and mitochondrion organization (GOterm4 and GOterm7). (F) Eleven out of 14 OCRGs in LUSC enriched in five significant pathways including mitochondrial fission, regulation of T cell activation, divalent metal ion transport, mitochondrial membrane organization, and mitochondrion organization. More OCRGs (TOMM40, APOO, PARL, HSPD1, MTFP1, TOMM22, OPA1, and MTFR2) enriched mitochondrion organization pathway (GOterm5). GO (Gene Ontology) chord was generated by http://www.bioinformatics.com.cn, an online platform for data analysis and visualization.
