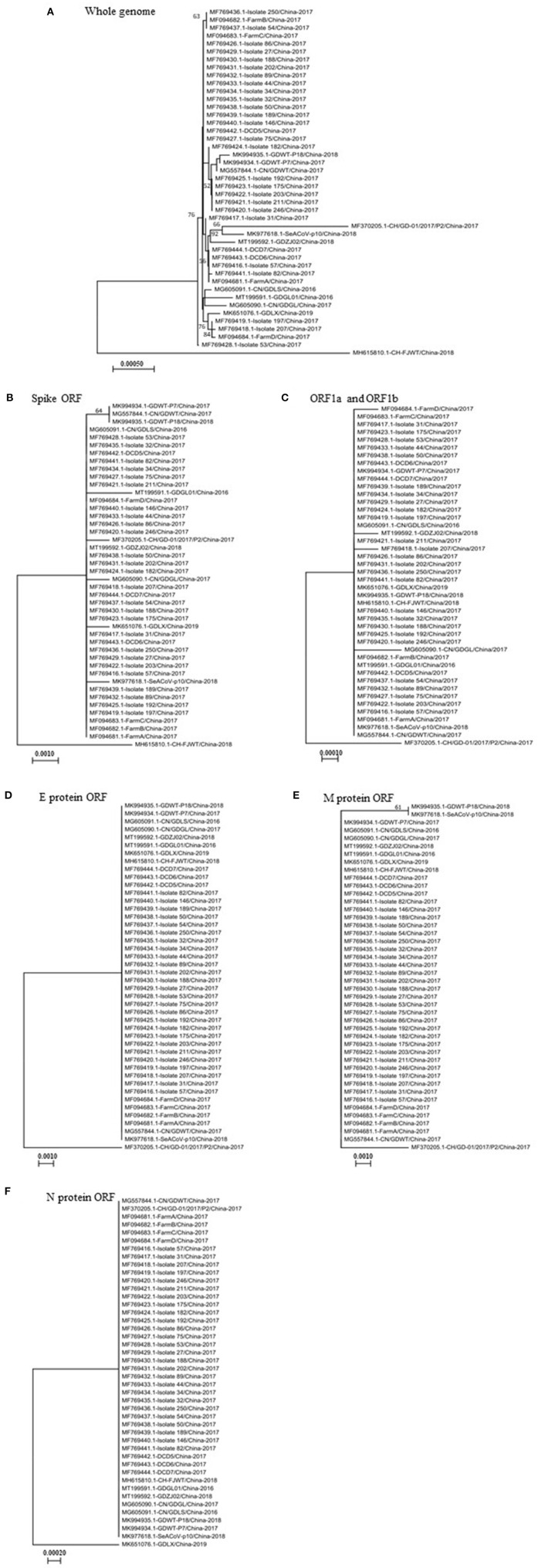
Figure 1.
The phylogenetic trees based on the whole genomes of SADS-CoVs (A) or the full-length spike glycoprotein ORF (B), ORF1a and ORF1b (C), or E protein ORF (D), M protein ORF (E), or N protein ORF (F). Multiple-sequence alignments were performed using Clustal Omega server and then phylogenetic trees were constructed using neighbor-joining method in the MEGA-X software (16, 17). The numbers at each branch represent bootstrap values >50% of 1,000 replicates. Scale bars indicate the number of inferred substitutions per site. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed using the Maximum Composite Likelihood method (17).