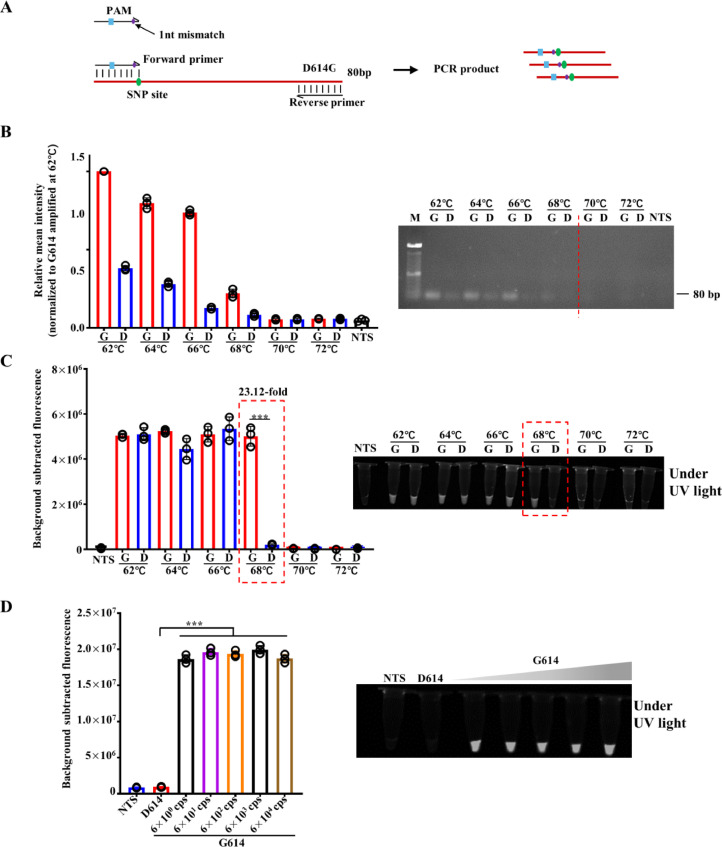
Fig. 3.
Validation of primer mismatch amplification of SARS-CoV-2 D614G by PCR and Cas12-based detection. (A) Schematic of the process for design of D614G specific primer with synthetic mismatch. The 3′ end of the forward primer is located at the SNP (A23403G) site of D614G mutation with a synthetic mismatch which is the same as that of crRNA-1. (B) Amplification of synthetic templates of D614 (6 × 104 cps) and G614 (6 × 104 cps) by mismatch primer designed in (A) annealed in a temperature gradient, respectively. After amplification, the products were analyzed by electrophoresis in 2% agarose gel and quantified. M represents 100 bp DNA ladder. D represents D614. G represents G614. The relative mean intensity was normalized to G614 amplified at 62°C. (C) Fluorescence signals of CRISPR/Cas12a-based detection of amplification products of (B) with crRNA-(-1). The presented images of visual detection were captured under UV light (n=3). (D) Fluorescence signals of D614 (6 × 104 cps) and 10-fold gradually diluted DNA templates of G614 (from 6 × 104 cps to 6 × 100 cps) detected by dsmCRISPR (n=3). NTS, non-target sequence. The presented images of visual detection were captured under UV light.