Key Points
Question
Is blood DNA methylation (DNAm) associated with the development of coronary heart disease (CHD)?
Findings
In this multi-cohort study, a high-dimensional multi-adjusted model assessing blood DNAm in 2321 American Indian adults selected 505 differentially methylated positions (DMPs) associated with incident CHD in the Strong Heart Study. These DMPs were evaluated in the Women’s Health Initiative, the Framingham Heart Study, and the Atherosclerosis Risk in Communities Study, and several DMPs common across cohorts tagged genes with established associations with cardiovascular disease.
Meaning
In this study, blood DNAm was associated with CHD beyond traditional factors associated with cardiovascular disease, with a complex epigenomic signature across populations.
This cohort study investigates the association of blood DNA methylation with incident coronary heart disease using a large number of methylation sites in a single model.
Abstract
Importance
American Indian communities experience a high burden of coronary heart disease (CHD). Strategies are needed to identify individuals at risk and implement preventive interventions.
Objective
To investigate the association of blood DNA methylation (DNAm) with incident CHD using a large number of methylation sites (cytosine-phosphate-guanine [CpG]) in a single model.
Design, Setting, and Participants
This prospective study, including a discovery cohort (the Strong Heart Study [SHS]) and 4 additional cohorts (the Women’s Health Initiative [WHI], the Framingham Heart Study [FHS], the Atherosclerosis Risk in Communities Study ([ARIC]–Black, and ARIC-White), evaluated 12 American Indian communities in 4 US states; African American women, Hispanic women, and White women throughout the US; White men and White women from Massachusetts; and Black men and women and White men and women from 4 US communities. A total of 2321 men and women (mean [SD] follow-up, 19.1 [9.2] years) were included in the SHS, 1874 women (mean [SD] follow-up, 15.8 [5.9] years) in the WHI, 2128 men and women (mean [SD] follow-up, 7.7 [1.8] years) in the FHS, 2114 men and women (mean [SD] follow-up, 20.9 [7.2] years) in the ARIC-Black, and 931 men and women (mean [SD] follow-up, 20.9 [7.2] years) in the ARIC-White. Data were collected from May 1989 to December 2018 and analyzed from February 2019 to May 2021.
Exposure
Blood DNA methylation.
Main Outcome and Measure
Using a high-dimensional time-to-event elastic-net model for the association of 407 224 CpG sites with incident CHD in the SHS (749 events), this study selected the differentially methylated CpG positions (DMPs) selected in the SHS and evaluated them in the WHI (531 events), FHS (143 events), ARIC-Black (350 events), and ARIC-White (121 events) cohorts.
Results
The median (IQR) age of participants in SHS was 55 (49-62) years, and 1359 participants (58.6%) were women. Elastic-net models selected 505 DMPs associated with incident CHD in the SHS beyond established risk factors, center, blood cell counts, and genetic principal components. Among those DMPs, 33 were commonly selected in 3 or 4 of the other cohorts and the pooled hazard ratios from the standard Cox models were significant at P < .05 for 10 of the DMPs. For example, the hazard ratio (95% CI) for CHD comparing the 90th and 10th percentiles of differentially methylated CpGs was 0.86 (0.78-0.95) for cg16604233 (tagged to COL11A2) and 1.23 (1.08-1.39) for cg09926486 (tagged to FRMD5). Some of the DMPs were consistent in the direction of the association; others showed associations in opposite directions across cohorts. Untargeted independent elastic-net models of CHD showed distinct DMPs, genes, and network of genes in the 5 cohorts.
Conclusions and Relevance
In this multi-cohort study, blood-based DNAm findings supported an association between a complex blood epigenomic signature and CHD that was largely different across populations.
Introduction
In the US, American Indian communities experience a disproportionate burden of coronary heart disease (CHD) compared with other racial and ethnic groups.1,2 Strategies are needed to identify individuals who are susceptible to CHD and to implement preventive interventions.2 Changes in epigenetic marks, in particular global DNA methylation (DNAm) and site-specific DNAm, are associated with differential risk of CHD and atherosclerosis.3,4,5,6 Extant studies have been limited by a lack of replication across populations, uncertain biological plausibility of the methylation sites identified (cytosine-phosphate-guanine [CpG]), and the assessment of differentially methylated CpG positions (DMPs) one by one or in nearby clusters without considering the complex interassociations across signals.
We investigated the association of DMPs and differentially methylated CpG regions (DMRs) from baseline blood DNA with the incidence of CHD in the Strong Heart Study (SHS), the largest and longest running study, to our best knowledge, of cardiovascular disease in American Indian communities.2,7 We conducted an untargeted epigenome-wide association study (EWAS) considering all CpGs simultaneously in a high-dimensional model. We then evaluated DMPs associated with CHD in the SHS in 4 other cohorts: the Women’s Health Initiative (WHI), Framingham Heart Study (FHS), and Atherosclerosis Risk in Communities Study (ARIC, analyzing Black participants and White participants separately). We also conducted an untargeted high-dimensional epigenome-wide approach in each of the 5 cohorts. In the SHS, our cohort of interest, we conducted classic EWAS analyzing DMPs and DMRs individually as well as a targeted analysis of DMPs associated with cardiovascular outcomes in previous studies.3,6
Methods
From 1989 to 1991, 4549 men and women aged 45 to 74 years who were members of 1 of 13 American Indian tribes from the Northern Plains (North and South Dakota), Southern Plains (Oklahoma), and Southwest (Arizona) consented to participate in the SHS. For the present study, 1 of the tribes declined, leaving 3517 potential participants. After applying eligibility criteria (eMethods and eFigure 1 in Supplement 1), a total of 2321 participants who were free of cardiovascular disease at baseline, not missing urinary metal data, and not missing data on factors associated with risk of cardiovascular disease were included (eTable 1 in Supplement 1). We compared the findings in SHS with data on blood DNAm and incident CHD from 1874 individuals in the WHI, 2128 individuals in the FHS, 2114 individuals in the ARIC-Black, and 931 individuals in the ARIC-White (eMethods in Supplement 1).
Protocols for collecting data in the SHS,7 WHI,8 FHS,9,10 and ARIC11 were similar across cohorts. Race and ethnicity were self-identified in all studies. In the SHS, all participants had a tribal affiliation with 1 of the 12 American Indian tribes participating in the study. In the ARIC and FHS, race and ethnicity were self-selected from the following options: Black/African American, White/Caucasian, American Indian/Alaska Native, and Asian/Pacific Islander. In the WHI, race and ethnicity were self-selected from the following options: American Indian/Alaskan Native, Asian/Pacific Islander, Black, Hispanic/Latina, White, and other. No specific analyses by race and ethnicity were conducted, although several cohorts included only 1 race or ethnicity. The studies were reviewed and approved by institutional review boards at their respective institutions. In addition, the SHS and the present study were approved by tribal research review boards and the Indian Health Service institutional review boards. All participants provided written informed consent.
Cardiovascular Incidence Follow-up
The end points were fatal and nonfatal CHD. The follow-up was from the time of blood drawn for DNAm measurement (1989 to 1991 for SHS, 1993 to 1998 for WHI, 2005 to 2008 for FHS, and 1990 to 1995 for ARIC) to the time of CHD event (2017 for SHS, 2016 for WHI, 2014 for FHS, and 2018 for ARIC). Follow-up was censored at the time of non-CHD death, loss to follow-up, or the last day of follow-up. The mean (SD) follow-up time among participants without a cardiovascular event was 19.1 (9.2) years in the SHS, 15.8 (5.9) years in the WHI, 7.7 (1.8) years in the FHS, 20.9 (7.2) years in the ARIC-Black, and 20.9 (7.2) years in the ARIC-White. All cohorts used a central adjudication system with 2 or more physicians to classify CHD events7,9,10 and included myocardial infarction and coronary deaths in their definition (eMethods in Supplement 1).
Microarray DNAm Measurement
Details of microarray DNAm measurements are in the eMethods in Supplement 1. In the SHS, bisulfite-converted blood DNA was measured using the MethylationEPIC BeadChip (Illumina 850K). We performed quality checks, data normalization, statistical preprocessing, and β-value calculations (methylation proportion at a given CpG), corrected batch effects, and estimated Houseman cell proportions (CD8T, CD4T, NK, B cells, monocytes, and granulocytes).12,13 The WHI, FHS, and ARIC used the Illumina Infinium HumanMethylation450K BeadChip array. After preprocessing, the number of CpGs was 788 368 in the SHS, 434 113 in the WHI, 408 254 in the FHS, 483 525 in the ARIC-Black, and 482 815 in the ARIC-White.
Statistical Analysis
Traditionally, the association of blood DNAm with health outcomes is analyzed in models for individual CpG sites. Accounting for the interassociations of CpG sites with the elastic-net tool might be more appropriate in addressing confounding and the complex associations across DMPs. We used GLMnet penalized regression (elastic-net) applied to survival time (R package glmnet [the R Foundation]) with α = .05 to account for complex interassociations and with the regularization parameter λ14 to achieve the minimum mean squared error, selected using 10-fold cross-validation (eMethods in Supplement 1). Our primary analysis was in the SHS. We compared DMPs selected in an elastic-net model with factors traditionally associated with risk and center (Table 1) with the elastic-net model further adding the 407 224 CpGs available from the 450 000 array, as well as blood cell counts and 5 genetic principle components (PCs). We annotated the DMPs to the nearest gene.15 To assess the relevance of the DMPs selected, we ran 2 sets of analyses in the other cohorts. First, we ran a targeted elastic-net model with the DMPs selected by the SHS in each of the other cohorts. Second, we ran untargeted elastic-net models in each of the other cohorts using all CpGs available, similar to our initial elastic-net SHS model. We also ran a targeted elastic-net model in the SHS and the 4 other cohorts introducing 248 DMPs that have been associated with incident CHD or atherosclerosis in previous studies3,6 to assess their replication in the populations in this study.
Table 1. Elastic-Net Models With Traditional Factors Associated With Risk of Coronary Heart Disease and the Same Models With Multiple Cytosine-Phosphate-Guanine Sites (CpGs).
| Modela | Parameters included in the model, No. | Parameters selected by the model, No. | ||
|---|---|---|---|---|
| Factors associated with risk | CpGs | Factors associated with risk | CpGs | |
| Strong Heart Study | ||||
| Model 1 | 11 | NA | 11 | NA |
| Model 2 | 21 | NA | 21 | NA |
| Model 2 and 450 000 array | 21 | 407 224 | 10 | 505 |
| Model 2 850 000 array | 21 | 788 368 | 10 | 635 |
| Model 2 and targeted CpGsb | 21 | 248 | 18 | 86 |
| Women’s Health Initiative | ||||
| Model 1 | 9 | NA | 9 | NA |
| Model 2 | 14 | NA | 10 | NA |
| Model 2 and 450 000 array | 14 | 434 113 | 6 | 398 |
| Model 2 and 505 CpGsc | 14 | 505 | 7 | 108 |
| Model 2 and targeted CpGsb | 14 | 248 | 11 | 56 |
| Framingham Heart Study | ||||
| Model 1 | 11 | NA | 9 | NA |
| Model 2 | 16 | NA | 14 | NA |
| Model 2 and 450 000 array | 16 | 408 254 | 2 | 698 |
| Model 2 and 505 CpGsc | 16 | 492 | 6 | 111 |
| Model 2 and targeted CpGsb | 16 | 245 | 9 | 73 |
| ARIC-Black | ||||
| Model 1 | 10 | NA | 10 | NA |
| Model 2 | 20 | NA | 19 | NA |
| Model 2 and 450 000 array | 20 | 483 525 | 7 | 1273 |
| Model 2 and 505 CpGsc | 20 | 503 | 13 | 219 |
| Model 2 and targeted CpGsb | 20 | 247 | 15 | 56 |
| ARIC-White | ||||
| Model 1 | 11 | NA | 10 | NA |
| Model 2 | 21 | NA | 19 | NA |
| Model 2 and 450 000 array | 21 | 482 815 | 4 | 819 |
| Model 2 + and 505 CpGsc | 21 | 503 | 12 | 109 |
| Model 2 and targeted CpGsb | 21 | 248 | 15 | 146 |
Abbreviations: ARIC, Atherosclerosis Risk in Communities Study; FHS, Framingham Heart Study; NA, not applicable; SHS, Strong Heart Study; WHI, Women’s Health Initiative.
Model 1 included traditional factors associated with risk, including age; smoking status (never, former, current); body mass index (calculated as weight in kilograms divided by height in meters squared); low-density lipoprotein cholesterol; high-density lipoprotein cholesterol; hypertension treatment (yes/no); type 2 diabetes (yes/no); and systolic blood pressure. All studies except the WHI further included sex. The SHS and the FHS further included albuminuria status (microalbuminuria, normal, macroalbuminuria). SHS and ARIC further included study center. FHS further included pedigree number. WHI further included race. Model 2 further included genetic principal components and blood cell counts for SHS and ARIC, only blood cell counts for WHI, and cell counts and plate number for FHS. ARIC further included 5 methylation batch effect principal components calculated using functional normalization.
This model includes 248 CpGs associated with atherosclerotic cardiovascular disease in previous studies.
CpGs included in the 450 000, selected by the elastic-net model using 450 000 data in the SHS (n = 505), and available in WHI, FHS, and ARIC. In some studies, some CpGs were not available and the number is lower than 505.
Elastic-net is an excellent tool to select DMPs associated with disease. However, it does not allow comparing effect estimates across studies. For DMPs selected by elastic-net in all the cohorts or in at least in 4 of the 5 cohorts, we calculated individual hazard ratios (HRs) and 95% CIs for their association with incident CHD using Cox regression adjusted as the elastic-net models. We then pooled the HRs for the common DMPs across cohorts using a random-effects meta-analysis (R package meta [the R Foundation]).16 In a sensitivity analysis, we restricted the follow-up to 10 years for the SHS (1989 to 1999) and ran elastic-net models in the FHS, WHI, and ARIC with the same follow-up for the SHS-selected DMPs.
In the SHS, we ran traditional EWAS using Cox regression for individual CpGs using all CpGs in the 850 000 array (untargeted analysis) and using DMPs associated with cardiovascular disease in previous studies (targeted analysis).3,6 We also tested differential methylation at the regional level using DMRcate. DMRs were annotated to the closest gene.17 All tests were 2-tailed, and significance was set at P < .05, accounting for multiple comparisons using the false discovery test (FDR) when indicated.
Protein-Protein Interaction Network
From the 450 000 DMPs reported in the SHS elastic-net model, we created a list of unique protein-coding genes. Protein interaction data were obtained from the STRING database v11.0,18 which estimates the likelihood the annotated interaction between a pair of proteins is biologically meaningful and reproducible. The network was analyzed using Cytoscape version 3.7.1 (Cytoscape Consortium).19 We kept connections obtained from experimental studies, public databases, and text mining with a minimum confidence score of 0.5. Unconnected nodes were excluded. We indicated which of the protein-coding genes were replicated in the targeted elastic-net model in the other cohorts. We also ran networks with the protein-coding genes identified by the untargeted elastic-net models in each cohort, and a network with the common protein-coding genes across all cohorts.
Results
The median (IQR) age of participants in SHS was 55 (49-62) years, and 1359 participants (58.6%) were women. A total of 749 participants (383 women) developed incident CHD over a mean (SD) of 11.7 (9.2 person-years of follow-up). Traditional factors associated with risk of CHD were observed (eTable 2 in Supplement 1).
Elastic-Net Model in the SHS and Replication in Other Cohorts
The elastic-net model, including 407 224 CpGs in the SHS together with factors traditionally associated with risk, blood cell counts, and genetic PCs, selected 505 DMPs associated with incident CHD (Table 1; eTable in Supplement 2). In the standard model, all factors associated with risk remained; in the model with the 505 DMPs, smoking and body mass index (BMI, calculated as weight in kilograms divided by height in meters squared) were no longer selected.
In elastic-net models implemented with these 505 DMPs, 108 were selected as associated with incident CHD in the WHI, 111 in the FHS, 219 in the ARIC-Black, and 109 in the ARIC-White. Four DMPs were common in all cohorts (cg02628823 [annotated to MGAT4D], cg04250451 [RUNX3], cg08178991 [ITGAX], cg16604233 [COL11A2]), and 29 were common in at least 4 cohorts. The pooled HRs from the standard Cox models were significant at P < .05 for 10 of these 33 DMPs (eg, the hazard ratio [95% CI] for CHD comparing the 90th and 10th percentiles of differentially methylated CpGs was 0.86 [0.78-0.95] for cg16604233 [tagged to COL11A2] and 1.23 [1.08-1.39] for cg09926486 [tagged to FRMD5]) (Table 2). These 10 DMPs generally showed HRs in the same direction of the association (inverse or positive) in all cohorts. The pooled HRs for the remaining 23 DMPs were not significant at P < .05, although some were borderline (eTable 3 in Supplement 1). Several HRs, including those for cg02628823 (MGAT4D), cg04250451 (RUNX3), and cg08178991 (ITGAX), showed inconsistent association across cohorts.
Table 2. Hazard Ratios (HRs) and 95% CIs of Coronary Heart Disease by Differentially Methylated Cytosine-Phosphate-Guanine Sites (CpGs) Initially Identified in the Strong Heart Study (SHS).
| CpGa | Chr | Gene | HR (95% CI)b | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| SHS | WHI | FHS | ARIC-Black | ARIC-White | Pooled HR (95% CI) | P value | |||
| cg01620164 | 2 | FIGN | 0.89 (0.69-1.13) | 0.90 (0.70-1.16) | 1.17 (0.63-2.16) | 0.60 (0.410.87) | 0.66 (0.35-1.21) | 0.82 (0.68-0.98) | .03 |
| cg12479512 | 3 | RBSN | 0.76 (0.64-0.91) | 0.86 (0.69-1.06) | 0.95 (0.61-1.49) | 0.67 (0.53-0.83) | 0.90 (0.78-1.03) | 0.85 (0.75-0.95) | .007 |
| cg16604233 | 6 | COL11A2 c | 0.89 (0.74-1.06) | 0.86 (0.67-1.10) | 0.54 (0.29-1.00) | 0.83 (0.65-1.07) | 0.71 (0.46-1.10) | 0.86 (0.78-0.95) | .002 |
| cg07964553 | 4 | NEUROG2 | 1.14 (1.05-1.25) | 1.18 (1.01-1.38) | 1.18 (0.92-1.51) | 1.11 (0.91-1.36) | 1.21 (0.92-1.60) | 1.11 (1.06-1.17) | <.001 |
| cg22293458 | 3 | VPS8 | 1.22 (1.06-1.40) | 1.09 (0.88-1.34) | 0.82 (0.49-1.38) | 1.17 (0.99-1.38) | 1.38 (0.96-1.97) | 1.11 (1.02-1.21) | .01 |
| cg26955383 | 10 | CALHM1 | 1.22 (1.01-1.47) | 1.12 (0.91-1.38) | 1.26 (0.81-1.95) | 1.13 (0.91-1.41) | 1.31 (0.71-2.42) | 1.17 (1.05-1.30) | .006 |
| cg02628823 | 4 | MGAT4D c | 1.23 (1.06-1.43) | 0.92 (0.74-1.15) | 1.21 (0.94-1.57) | 1.11 (0.91-1.35) | 1.60 (1.22-2.11) | 1.17 (1.01-1.36) | .04 |
| cg01297357 | 12 | ASIC1 | 1.27 (1.08-1.49) | 1.02 (0.83-1.26) | 1.17 (0.87-1.57) | 1.61 (1.25-2.08) | 1.50 (0.88-2.54) | 1.21 (1.04-1.40) | .01 |
| cg09926486 | 15 | FRMD5 | 1.29 (1.08-1.53) | 1.11 (0.90-1.38) | 0.88 (0.56-1.39) | 1.39 (1.12-1.73) | 1.24 (0.84-1.82) | 1.23 (1.08-1.39) | .001 |
| cg08622677 | 12 | PRMT8 | 1.25 (1.03-1.51) | 1.06 (0.86-1.32) | 1.23 (0.77-1.97) | 1.32 (1.07-1.63) | 1.76 (1.10-2.82) | 1.23 (1.07-1.42) | .005 |
Abbreviations: ARIC, the Atherosclerosis Risk in Communities Study; Chr, chromosome; FHS, the Framingham Heart Study; WHI, the Women’s Health Initiative.
The CpGs are ordered based on the pooled HRs.
HRs and 95% CIs comparing the 90th and 10th percentiles of differentially methylated CpGs. HRs correspond to those estimated by Cox regression with that CpG entered in the model together with traditional factors associated with risk but without adjustment for other CpGs. Model 1 included traditional factors associated with risk, including age; smoking status (never, former, current); body mass index (calculated as weight in kilograms divided by height in meters squared); low-density lipoprotein cholesterol; high-density lipoprotein cholesterol; hypertension treatment (yes/no); type 2 diabetes (yes/no); and systolic blood pressure. All studies except the WHI further included sex. The SHS and the FHS further included albuminuria status (microalbuminuria, normal, macroalbuminuria). SHS and ARIC further included study center. FHS further included pedigree number. WHI further included race. Model 2 further included genetic principle components and blood cell counts for SHS and ARIC, only blood cell counts for WHI, and cell counts and plate number for FHS. ARIC further included 5 methylation batch effect principle components calculated using functional normalization.
These differentially methylated positions were selected as associated with incident coronary heart disease in the untargeted elastic-net in SHS and subsequently by targeted elastic-net models in the other 4 cohorts; all other differentially methylated positions were selected in SHS and in 3 other cohorts.
In the protein-protein interaction analysis, 466 unique genes associated with the 505 DMPs identified by the SHS elastic-net models were included. We discarded 235 noncoding RNA genes or unconnected nodes. From 231 nodes in the network (Figure), 3 were common for the 5 cohorts, 14 were common for 4 cohorts, 66 were common for 3 cohorts, 78 were common for the SHS with another cohort (18 with FHS, 9 with WHI, 43 with ARIC-Black, and 8 with ARIC-White), and 70 were selected by SHS only. The EGFR gene was a hub with 35 interactions, followed by SMAD3 with 13 interactions and ADCY3 with 12 interactions.
Figure. Protein-Protein Interaction Network of Genes Annotated to Differentially Methylated Positions (DMPs) Initially Associated With Incident Coronary Heart Disease in the Strong Heart Study (SHS) and Subsequently Implemented in 4 Other Cohorts.
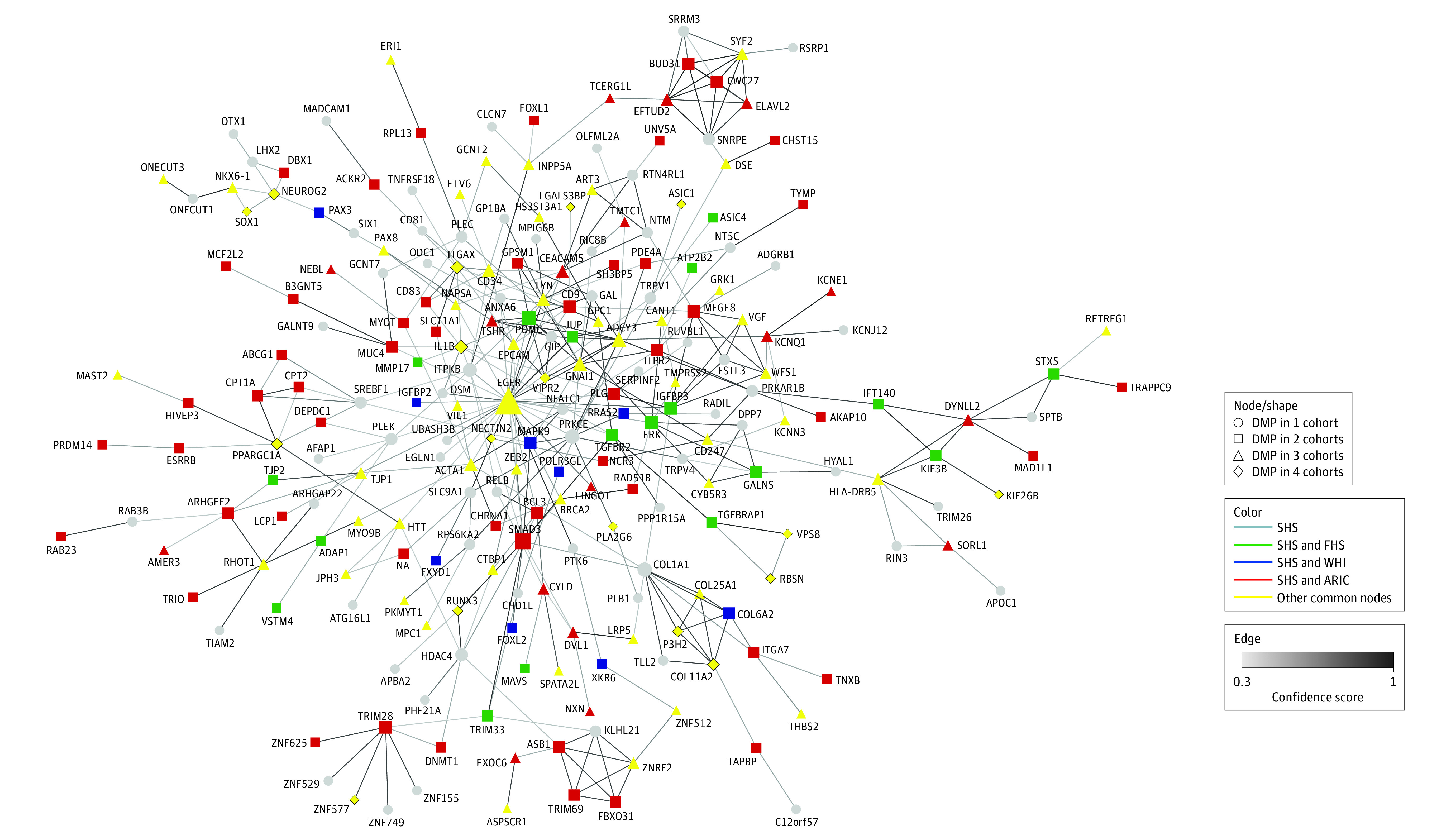
The network includes 231 nodes. The gray nodes indicate DMPs in the SHS only; green, the SHS and the Framingham Heart Study (FHS); blue, the SHS and the Women’s Health Initiative (WHI); red, the SHS and either both the Atherosclerosis Risk in Communities Study [ARIC]–Black and ARIC-White (triangle) or only 1 of the ARIC cohorts (square); yellow, other nodes of 3 or more cohorts. Diamonds correspond to the DMPs identified in the untargeted analysis and common across the 5 cohorts. The size of the nodes is proportional to the number of connections. The edges indicate confidence score interaction (only confidence scores ≥0.5 were included).
Independent Untargeted Elastic-Net Models in All Cohorts
In the untargeted elastic-net models implemented with all 450 000 CpGs available in each cohort, 188 DMPs were selected as associated with incident CHD in the WHI, 235 in the FHS, 1273 in the ARIC-Black, and 819 in the ARIC-White (eTable in Supplement 2), in addition to the 505 DMPs previously described for SHS. Running ad hoc targeted elastic-net models with the DMPs selected in each of the other cohorts resulted in 141 DMPs selected by 4 or 5 cohorts, including 14 selected by all 5 (Table 3). When we estimated the pooled HRs for those 141 DMPs using Cox models, 35 were significant at P < .05, most of them with consistent direction of the association across cohorts, including CpGs annotated to AHRR and MTHFR (HR [95% CI] comparing 90th to 10th percentile of differentially methylated cg05575921, 0.68 [0.54-0.85]; for cg05228408, 0.77 [0.65-0.91]) (eTable 4 in Supplement 1).
Table 3. Hazard Ratios (HRs) and 95% CIs of Coronary Heart Disease by Differentially Methylated Cytosine-Phosphate-Guanine Sites (CpGs) Initially Selected in All 5 Cohorts.
| CpGa | Chr | Gene | HR (95% CI)b | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| SHS | WHI | FHS | ARIC-Black | ARIC-White | Pooled HR (95% CI) | P value | |||
| cg03068497 | 7 | GARS1 | 0.90 (0.74-1.11) | 0.84 (0.66-1.06) | 0.58 (0.34-0.99) | 0.83 (0.63-1.09) | 0.45 (0.28-0.75) | 0.70 (0.53-0.93) | .01 |
| cg05228408 | 1 | MTHFR | 0.88 (0.72-1.09) | 0.84 (0.67-1.06) | 0.59 (0.33-1.06) | 0.67 (0.50-0.91) | 0.63 (0.39-1.03) | 0.77 (0.65-0.91) | .003 |
| cg23933602 | 10 | RSU1 | 0.84 (0.70-1.00) | 0.73 (0.57-0.95) | 0.44 (0.23-0.84) | 0.79 (0.65-0.96) | 0.73 (0.43-1.22) | 0.81 (0.74-0.89) | E-05 |
| cg05823563 | 9 | C9orf125 | 1.08 (0.9-1.29) | 0.87 (0.69-1.1) | 0.37 (0.21-0.66) | 1.06 (0.79-1.42) | 0.72 (0.54-0.97) | 0.83 (0.64-1.07) | .16 |
| cg16604233 | 6 | COL11A2 | 0.89 (0.74-1.06) | 0.86 (0.67-1.10) | 0.54 (0.29-1.00) | 0.83 (0.65-1.07) | 0.71 (0.46-1.10) | 0.86 (0.78-0.95) | .002 |
| cg10574494 | 4 | SPATA18 | 1.12 (0.93-1.35) | 0.85 (0.68-1.08) | 0.40 (0.22-0.73) | 1.37 (1.07-1.76) | 0.68 (0.41-1.12) | 0.88 (0.64-1.22) | .45 |
| cg08447739 | 10 | HTRA1 | 0.89 (0.74-1.07) | 0.69 (0.54-0.88) | 1.28 (0.84-1.97) | 0.94 (0.72-1.23) | 0.84 (0.52-1.37) | 0.89 (0.77-1.03) | .11 |
| cg04250451 | 1 | RUNX3 | 1.38 (1.15-1.65) | 0.97 (0.88-1.07) | 0.66 (0.42-1.03) | 0.99 (0.73-1.34) | 1.88 (1.08-3.27) | 1.04 (0.95-1.15) | .39 |
| cg08178991 | 16 | ITGAX | 1.44 (1.2-1.74) | 1.10 (0.9-1.34) | 1.56 (0.89-2.72) | 0.86 (0.77-0.96) | 1.25 (0.75-2.07) | 1.07 (0.95-1.2) | .24 |
| cg02628823 | 4 | MGAT4D | 1.23 (1.06-1.43) | 0.92 (0.74-1.15) | 1.21 (0.94-1.57) | 1.11 (0.91-1.35) | 1.60 (1.22-2.11) | 1.17 (1.01-1.36) | .04 |
| cg22454769 | 2 | FHL2 | 1.00 (0.81-1.23) | 1.17 (0.89-1.53) | 1.73 (0.90-3.33) | 1.41 (1.04-1.92) | 1.24 (0.73-2.12) | 1.17 (1.01-1.36) | .04 |
| cg05190790 | 11 | SOX1 | 1.05 (0.89-1.25) | 1.29 (1.02-1.63) | 1.27 (0.78-2.09) | 1.58 (1.19-2.10) | 1.23 (0.77-1.95) | 1.26 (1.06-1.49) | .008 |
| cg09476997 | 16 | SLC9A3R2 | 1.39 (1.13-1.70) | 1.16 (0.92-1.45) | 1.93 (1.05-3.54) | 1.41 (1.03-1.93) | 1.56 (0.96-2.52) | 1.28 (1.14-1.44) | 2.8E-05 |
| cg13543355 | 14 | LRRC74A | 1.13 (0.95-1.35) | 1.17 (0.95-1.44) | 1.16 (0.74-1.83) | 1.97 (1.60-2.43) | 1.94 (1.01-3.73) | 1.28 (1.05-1.57) | .02 |
Abbreviations: ARIC, the Atherosclerosis Risk in Communities Study; Chr, chromosome; FHS, the Framingham Heart Study; SHS, the Strong Heart Study; WHI, the Women’s Health Initiative.
The CpGs are ordered based on the pooled HRs.
HRs correspond to those estimated by Cox regression with that CpG entered in the model together with traditional risk factors but without adjustment for other CpGs and ordered based on the pooled HR in the meta-analysis. HRs and 95% CIs comparing the 90th and 10th percentiles of differentially methylated CpGs. HRs correspond to those estimated by Cox regression with that CpG entered in the model together with traditional factors associated with risk but without adjustment for other CpGs. Model 1 included traditional factors associated with risk, including age; smoking status (never, former, current); body mass index (calculated as weight in kilograms divided by height in meters squared); low-density lipoprotein cholesterol; high-density lipoprotein cholesterol; hypertension treatment (yes/no); type 2 diabetes (yes/no); and systolic blood pressure. All studies except the Women’s Health Initiative (WHI) further included sex. The Strong Heart Study (SHS) and the Framingham Heart Study (FHS) further included albuminuria status (microalbuminuria, normal, macroalbuminuria). SHS and ARIC further included study center. FHS further included pedigree number. WHI further included race. Model 2 further included genetic principle components and blood cell counts for SHS and ARIC, only blood cell counts for WHI, and cell counts and plate number for FHS. ARIC further included 5 methylation batch effect principle components calculated using functional normalization.
In the protein-protein interaction analysis for these untargeted elastic-net models, each cohort showed a largely distinct network of genes, with the number of nodes (most-connected gene) being 231 (EGFR) in the SHS, 133 (MAPK8) in the WHI, 293 (FN1) in the FHS, 732 (SRC) in the ARIC-Black, and 441 (CDC42) in the ARIC-White. The network of commonly annotated genes across 2 or more cohorts had 139 nodes (eFigure 2 in Supplement 1). The most connected node (selected by SHS, FHS, ARIC-Black, and ARIC-White) was HDAC4, a histone deacetylase with strong evidence that it regulates vascular inflammation via autophagy.20
Targeted Models With DMPs From Previous Studies
In a targeted analysis of 248 DMPs associated with cardiovascular disease in previous studies, 16 were associated with incident CHD in the SHS at P < .05 (eTable 5 in Supplement 1), none of them included in the 505 DMPs selected by elastic-net in the SHS. Using these 248 DMPs in targeted elastic-net models in each cohort, the number of DMPs associated with CHD ranged from 56 in the WHI and ARIC-Black to 146 in the ARIC-White (Table 1).
Traditional EWAS With CpGs Analyzed Individually (DMPs) or in Nearby Regions (DMRs)
In an untargeted EWAS with 850 000 CpG analyzed individually in the SHS, no DMPs were associated with incident CHD at FDR P < .05, 11 DMPs showed FDR P < .10 (eg, the HR [95% CI] for cg10812236 [tagged to PLEK] was 2.24 [1.66-3.02]) (eTable 6 in Supplement 1), and 69 866 DMPs showed P < .05. Five of the 11 DMPs were present in the 450 000, but none were associated with incident CHD in the other cohorts. The most frequent gene among the top untargeted DMPs in the SHS was PLEK (pleckstrin). Four DMPs in PLEK had FDR P < .10 (eFigure 3 in Supplement 1); of these, the untargeted SHS elastic-net model selected cg04872689. In the protein-protein network, a PLEK node was found with 6 interactions (Figure).
No DMRs passed the Stouffer P < .05 cutoff in the SHS, although 1 was .05 and 4 ranged from .08 to .09 (eTable 7 in Supplement 1). The top DMR was annotated to LINGO1. A DMP annotated to this gene was selected by the SHS elastic-net model and remained selected by ARIC-Black and ARIC-White in the targeted elastic-net. LINGO1 was also a node linked to EGFR in the protein-protein interaction network. Another LINGO1-annotated DMP was selected in the untargeted ARIC-White elastic-net.
Sensitivity Analysis by Length of Follow-up
The SHS elastic-net model, including all 450 000 CpGs restricted to 10-year follow-up, selected 625 DMPs. Introducing these DMPs in the other cohorts with a 10-year follow-up selected 99 DMPs for WHI, 122 for FHS, 266 for ARIC-Black, and 188 for ARIC-White.
Discussion
This high-dimensional EWAS of blood DNA methylation in American Indian adults identified a complex blood epigenomic signature associated with CHD, including shared DMPs in other populations. From more than 400 000 CpGs introduced with risk factors traditionally associated with CHD in the SHS, elastic-net models selected 505 DMPs associated with incident CHD. When these DMPs were modeled in the WHI, FHS, ARIC-Black, and ARIC-White using elastic-net, 33 DMPs were selected in 3 or 4 cohorts, some of them with consistent direction of association across cohorts and others inconsistent. In protein-protein interaction analyses, many genes were connected with a large hub for EGFR based on SHS, FHS, and ARIC-Black signals and with a hub for ITGAX based on the 5 cohorts. The targeted protein-protein network analysis supported overlap in interconnected biological pathways among the cohorts despite multiple DMPs unique to the SHS (eg, PLEK). The complex blood epigenomic signature of cardiovascular disease across populations was further revealed by the distinct DMPs, genes, and network of genes identified by untargeted elastic-net models of CHD in the 5 cohorts, although ad hoc targeted analyses suggest it is possible to identify common DMPs relevant for multiple populations.
Among the 33 DMPs common in 4 or 5 cohorts, the pooled HR was significant for 10, and most HRs were in the same direction across cohorts. Several of these DMPs were annotated to genes with a possible association with CHD. COL11A2 (collagen type XI α 2 chain) encodes a constituent of the extracellular matrix, and SNPs have been associated with coronary artery lesions.21 Several collagen-related DMPs were nodes in the protein-network analyses. MGAT4D is involved in the biosynthesis of n-glycan, a biomarker of cardiometabolic risk,22 and SNPs have been shown to be associated with blood pressure.23 ASIC1 is involved in calcium activation in vascular smooth muscle cells and has been shown to be associated with pulmonary hypertension and stroke.24,25 FRMD5 (FERN-kinase domain containing 5) SNPs are associated with lipids levels,26,27 and FERN is involved in cell adhesion.28 Less is known about cardiovascular associations with genes such as NEUROG2, encoding a transcription factor expressed in neural progenitor cells,29 or PRMT8, a methyltransferase expressed in the brain30 but with SNPs associated with lipids and CHD.31
Among 23 DMPs selected as associated with CHD in 4 or 5 cohorts but with nonsignificant pooled HRs, DMPs were positive in some but inverse in others, except for those annotated to PLA2G6, MMEL1, and GPR155, which were inverse in all cohorts and borderline significant. Many of the inconsistent DMPs were annotated to cardiovascular disease-relevant genes, such as IL1B, encoding interleukin-1β, a cytokine associated with atherosclerosis in the vasculature and systemically.32,33,34,35 In the protein network, IL1B interacted with ITGAX, which encodes a fibrinogen receptor that mediates cell-cell interactions and has been associated with hyperlipidemia and atherosclerosis.36,37 Other genes included SPPL2B, encoding a protease that cleaves the intracellular domain of tumor necrosis factor α, with roles in inflammation, cell survival, growth, and apoptosis38,39; RUNX3, involved in endothelial-mesenchymal transition and endothelial function40,41; PPRGC1A, encoding peroxisome proliferator-activated receptor γ coactivator 1-α, involved in energy and metabolism processes, including cardiac metabolism42,43; and PCDHGA4, part of the protocadherin γ gene subcluster. Exposure to diabetes in utero has been associated with PCDHGA4 hypomethylation in 388 Pima Indian People44 in one of the few epigenetic studies in American Indian populations beyond the SHS.
It is unclear why some DMPs show different directions in the association with CHD across cohorts. The biological plausibility for the DMPs annotated to IL1B, RUNX3, ITGAX, and PPRGC1A is even stronger than for the DMPs with consistent associations across cohorts. This inconsistency, also observed in previous EWAS,3,6 could be related to the plasticity and adaptability of epigenetic response during pathophysiological processes,45 and to differences in times of assessment, treatments, other factors associated with risk, and complex signatures and dynamics across cohorts. If these inconsistent DMPs reflect a biologically relevant phenomenon, pooling HRs across cohorts, as is traditionally done in EWAS, would miss many relevant DMPs. The inconsistencies could also reflect confounded associations by methylation in other DMPs not included in Cox models. Elastic-net coefficients, however, cannot be compared across cohorts as the model forces less-associated variables to have zero coefficients. Indeed, elastic-net allows identifying common DMPs associated with CHD across populations, but not comparing the effect estimates.
Some of the methylation sites associated with CHD might not be independent of traditional factors associated with risk. DNAm is strongly associated with obesity and smoking, including in the SHS.46,47 For smoking, DNAm sites might be more objective than self-reports. For example, cg1439173, associated with smoking and annotated to PRSS23, was selected by the SHS elastic-net as associated with CHD. The elastic-net model with the 505 CpGs no longer included smoking and obesity in the SHS, smoking in the FHS and ARIC-Black, or BMI in the WHI and ARIC-White.
EWAS of clinical cardiovascular outcomes have been challenging, which is expected to be because of the intricate pathophysiology of cardiovascular outcomes, including inflammation, oxidative stress, endothelial function, and thrombosis, among other pathways, and the numerous cells and tissues involved, including systemic effects. While our untargeted elastic-net models across cohorts provide a glimpse of these complexities, the common DMPs identified by the ad hoc targeted elastic-net models and protein-protein interaction network may contribute to identifying relevant signatures across populations. We identified a substantial number of consistent DMPs in subsequent Cox models. These DMPs included cg05575921 (annotated to AHRR), a smoking related DMP associated with cardiovascular disease in previous studies,48,49,50,51 and cg05228408 (annotated to MTHFR, involved in the regulation of homocysteine and with SNPs possibly associated with cardiovascular risk).52 Additional research can evaluate the usefulness of these DMPs for CHD and of the untargeted-targeted elastic-net strategy for identifying additional relevant DMPs in the future.
Our primary cohort was the SHS and our findings may be relevant for American Indian populations. American Indian individuals are at higher risk of CHD compared with other race and ethnicity groups in the US. There is need for improved identification of individuals at risk and for improved treatment and prevention strategies. We have focused this discussion on DMPs that overlap with WHI, FHS, and ARIC. Among DMPs that do not overlap, some may be of interest for American Indian individuals. Replication of these DMPs in other American Indian populations is needed, although there are currently few studies and their sample sizes are small.44,53
None of the DMPs selected by the elastic-net model coincided with the top signals in the regular EWAS in the SHS except for PLEK. This is possibly because of limited power, need to account for multiple comparisons, and lack of accounting for interassociations across CpGs in the traditional EWAS. Elastic-net minimizes multiple comparisons and genomic inflation, as one single model is run. Additionally, many of the DMPs selected by elastic-net had biological relevance, adding value to the findings and potential use. PLEK, for instance, encodes a protein involved in calcium signaling and it was differentially expressed in abdominal aortic aneurysms compared with aorta tissue from control individuals.54 Many DMPs were annotated to genes whose function is presently unknown. Experimental research in these genetic regions may contribute to discovering new pathways relevant for CHD.
Limitations
Our study has important limitations. Power was limited for some cohorts, and we could not replicate the findings in other American Indian populations. Using these data for risk prediction is premature given their complexity. For instance, assessing predictive ability is problematic as training a new model different from that of the discovery set might overestimate it in replication sets.55 Elastic-net does not allow modeling DMRs. We annotated each CpG to the nearest gene, but this annotation can be uncertain. Additionally, although the use of Cox regression following elastic-net to obtain hazard ratios for each DMP and pool across cohorts is not standard, we used this strategy to compare effect estimates across cohorts. Strengths of our study include the high-quality outcome ascertainment and quality control procedures in all the cohorts and the diverse race and ethnicity composition.
Conclusions
In this multi-cohort study, we found that the blood epigenomic signature of cardiovascular disease is complex and distinct across populations. However, key findings from the SHS were generalized to the WHI, FHS, and ARIC, indicating that DNAm may help identify individuals at higher risk of developing CHD if methods that account for high-dimensionality of the data are used. We identified DMPs with relevant biological associations with atherosclerosis. The different direction of the association across studies for some DMPs could be biologically relevant and needs to be further investigated. Our blood-based DNAm findings open opportunities for identifying individuals at risk of CHD beyond risk factors traditionally associated with cardiovascular disease.
eMethods
eTable 1. Descriptive characteristics for eligible Strong Heart Study participants versus finally selected participants
eTable 2. Strong Heart Study baseline participants’ characteristics by coronary heart disease (CHD) incidence status
eTable 3: Hazard ratios (95% CIs) of CpGs available in the Illumina 450 000 platform initially associated with CHD by elastic-net in the Strong Heart Study (SHS), subsequently associated in the Women’s Health Initiative (WHI), the Framingham Heart Study (FHS) and the Atherosclerosis Risk in Communities (ARIC) Study, and not statistically significant in the meta-analysis
eTable 4. Hazard ratios (95% confidence intervals) of incident coronary heart disease comparing the 90th vs 10th percentile of differentially methylated CpGs available in the Illumina 450 000 platform and selected by untargeted elastic-net in each of the study cohorts, replicated with targeted elastic-net models in at least three of the other cohorts, and statistically significant in the meta-analysis
eTable 5: Targeted approach for the association of differentially methylated CpGs associated with atherosclerotic cardiovascular disease in previous studies and their association with incident CHD in the Strong Heart Study
eTable 6: Untargeted approach (EWAS) for coronary heart disease in the Strong Heart Study and replication in the Framingham Heart Study and the Women’s Health Initiative
eTable 7: Differentially methylated regions (DMRs) associated with incident coronary heart disease (CHD) in the Strong Heart Study
eFigure 1. Flowchart of the data exclusion process of the Strong Heart Study
eFigure 2. Protein-protein interaction network of genes annotated to DMPs selected by untargeted elastic-net models in two or more cohorts
eFigure 3: Hazard ratio (95% confidence intervals) for coronary heart disease and several hypermethylated CpGs annotated to PLEK in the Strong Heart Study
eReferences
eTable. Differentially Methylated Positions (DMPs) selected by an untargeted elastic-net model in the Strong Heart Study using all 450 000 CpGs available
References
- 1.Howard BV, Lee ET, Cowan LD, et al. Rising tide of cardiovascular disease in American Indians. the Strong Heart Study. Circulation. 1999;99(18):2389-2395. doi: 10.1161/01.CIR.99.18.2389 [DOI] [PubMed] [Google Scholar]
- 2.Breathett K, Sims M, Gross M, et al. ; American Heart Association Council on Epidemiology and Prevention; Council on Quality of Care and Outcomes Research; Council on Cardiovascular and Stroke Nursing; Council on Clinical Cardiology; and Council on Lifestyle and Cardiometabolic Health . Cardiovascular health in American Indians and Alaska Natives: a scientific statement from the American Heart Association. Circulation. 2020;141(25):e948-e959. doi: 10.1161/CIR.0000000000000773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agha G, Mendelson MM, Ward-Caviness CK, et al. Blood leukocyte DNA methylation predicts risk of future myocardial infarction and coronary heart disease. Circulation. 2019;140(8):645-657. doi: 10.1161/CIRCULATIONAHA.118.039357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donahue JK. Editorial commentary: epigenetics and cardiovascular disease—from concept to reality. Trends Cardiovasc Med. 2018;28(5):320-321. doi: 10.1016/j.tcm.2018.02.005 [DOI] [PubMed] [Google Scholar]
- 5.van der Harst P, de Windt LJ, Chambers JC. Translational perspective on epigenetics in cardiovascular disease. J Am Coll Cardiol. 2017;70(5):590-606. doi: 10.1016/j.jacc.2017.05.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Sanlés A, Sayols-Baixeras S, Subirana I, Degano IR, Elosua R. Association between DNA methylation and coronary heart disease or other atherosclerotic events: a systematic review. Atherosclerosis. 2017;263:325-333. doi: 10.1016/j.atherosclerosis.2017.05.022 [DOI] [PubMed] [Google Scholar]
- 7.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study. a study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132(6):1141-1155. doi: 10.1093/oxfordjournals.aje.a115757 [DOI] [PubMed] [Google Scholar]
- 8.Manson JE, Chlebowski RT, Stefanick ML, et al. Menopausal hormone therapy and health outcomes during the intervention and extended poststopping phases of the Women’s Health Initiative randomized trials. JAMA. 2013;310(13):1353-1368. doi: 10.1001/jama.2013.278040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curb JD, McTiernan A, Heckbert SR, et al. ; WHI Morbidity and Mortality Committee . Outcomes ascertainment and adjudication methods in the Women’s Health Initiative. Ann Epidemiol. 2003;13(9)(suppl):S122-S128. doi: 10.1016/S1047-2797(03)00048-6 [DOI] [PubMed] [Google Scholar]
- 10.D’Agostino RB Sr, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117(6):743-753. doi: 10.1161/CIRCULATIONAHA.107.699579 [DOI] [PubMed] [Google Scholar]
- 11.The ARIC investigators . The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129(4):687-702. doi: 10.1093/oxfordjournals.aje.a115184 [DOI] [PubMed] [Google Scholar]
- 12.Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558-560. doi: 10.1093/bioinformatics/btw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triche TJ Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastie T, Qian J, Tay K. An introduction to glmnet. Accessed July 4, 2021. https://web.stanford.edu/~hastie/glmnet/glmnet_alpha.html
- 15.Illumina Inc . Infinium MethylationEPIC product files. Accessed July 4, 2021. https://support.illumina.com/downloads/infinium-methylationepic-v1-0-product-files.html
- 16.Viechtbauer W. Metafor: meta-analysis package for R. Accessed April 1, 2021. https://cran.r-project.org/web/packages/metafor/metafor.pdf
- 17.Hansen K. IlluminaHumanMethylationEPICanno.ilm10b2.hg19: annotation for Illumina's EPIC methylation arrays.1 R package version 0.6.0. Accessed July 4, 2021. https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylationEPICmanifest.html
- 18.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D, Xiao C, Long F, et al. HDAC4 regulates vascular inflammation via activation of autophagy. Cardiovasc Res. 2018;114(7):1016-1028. doi: 10.1093/cvr/cvy051 [DOI] [PubMed] [Google Scholar]
- 21.Sheu JJ, Lin YJ, Chang JS, et al. Association of COL11A2 polymorphism with susceptibility to Kawasaki disease and development of coronary artery lesions. Int J Immunogenet. 2010;37(6):487-492. doi: 10.1111/j.1744-313X.2010.00952.x [DOI] [PubMed] [Google Scholar]
- 22.Wittenbecher C, Štambuk T, Kuxhaus O, et al. Plasma N-glycans as emerging biomarkers of cardiometabolic risk: a prospective investigation in the EPIC-Potsdam Cohort Study. Diabetes Care. 2020;43(3):661-668. doi: 10.2337/dc19-1507 [DOI] [PubMed] [Google Scholar]
- 23.Li C, He J, Chen J, et al. Genome-wide gene-sodium interaction analyses on blood pressure: the Genetic Epidemiology Network of Salt-Sensitivity Study. Hypertension. 2016;68(2):348-355. doi: 10.1161/HYPERTENSIONAHA.115.06765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert LM, Resta TC, Jernigan NL. RhoA increases ASIC1a plasma membrane localization and calcium influx in pulmonary arterial smooth muscle cells following chronic hypoxia. Am J Physiol Cell Physiol. 2018;314(2):C166-C176. doi: 10.1152/ajpcell.00159.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiang M, Dong X, Zha Z, et al. Selection of an ASIC1a-blocking combinatorial antibody that protects cells from ischemic death. Proc Natl Acad Sci U S A. 2018;115(32):E7469-E7477. doi: 10.1073/pnas.1807233115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Yin RX, Zhang QH, et al. Association of the FRMD5 rs2929282 polymorphism and serum lipid profiles in two Chinese ethnic groups. Int J Clin Exp Pathol. 2018;11(7):3494-3510. [PMC free article] [PubMed] [Google Scholar]
- 27.Baeyens N, Latrache I, Yerna X, Noppe G, Horman S, Morel N. Redundant control of migration and adhesion by ERM proteins in vascular smooth muscle cells. Biochem Biophys Res Commun. 2013;441(3):579-585. doi: 10.1016/j.bbrc.2013.10.118 [DOI] [PubMed] [Google Scholar]
- 28.Seo Y, Park J, Choi W, et al. Antiatherogenic effect of resveratrol attributed to decreased expression of ICAM-1 (intercellular adhesion molecule-1). Arterioscler Thromb Vasc Biol. 2019;39(4):675-684. doi: 10.1161/ATVBAHA.118.312201 [DOI] [PubMed] [Google Scholar]
- 29.Deng B, Gou X, Chen H, et al. Targeted delivery of neurogenin-2 protein in the treatment for cerebral ischemia-reperfusion injury. Biomaterials. 2013;34(34):8786-8797. doi: 10.1016/j.biomaterials.2013.07.076 [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280(38):32890-32896. doi: 10.1074/jbc.M506944200 [DOI] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Ripatti S, Lindqvist I, et al. ; ENGAGE Consortium . Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47-55. doi: 10.1038/ng.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70(18):2278-2289. doi: 10.1016/j.jacc.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viana-Huete V, Fuster JJ. Potential therapeutic value of interleukin 1β-targeted strategies in atherosclerotic cardiovascular disease. Rev Esp Cardiol (Engl Ed). 2019;72(9):760-766. doi: 10.1016/j.recesp.2019.02.021 [DOI] [PubMed] [Google Scholar]
- 34.Pedersen BK. Anti-inflammatory effects of exercise: role in diabetes and cardiovascular disease. Eur J Clin Invest. 2017;47(8):600-611. doi: 10.1111/eci.12781 [DOI] [PubMed] [Google Scholar]
- 35.Everett BM, MacFadyen JG, Thuren T, Libby P, Glynn RJ, Ridker PM. Inhibition of interleukin-1β and reduction in atherothrombotic cardiovascular events in the CANTOS trial. J Am Coll Cardiol. 2020;76(14):1660-1670. doi: 10.1016/j.jacc.2020.08.011 [DOI] [PubMed] [Google Scholar]
- 36.Gower RM, Wu H, Foster GA, et al. CD11c/CD18 expression is upregulated on blood monocytes during hypertriglyceridemia and enhances adhesion to vascular cell adhesion molecule-1. Arterioscler Thromb Vasc Biol. 2011;31(1):160-166. doi: 10.1161/ATVBAHA.110.215434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Gower RM, Wang H, et al. Functional role of CD11c+ monocytes in atherogenesis associated with hypercholesterolemia. Circulation. 2009;119(20):2708-2717. doi: 10.1161/CIRCULATIONAHA.108.823740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fluhrer R, Grammer G, Israel L, et al. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nat Cell Biol. 2006;8(8):894-896. doi: 10.1038/ncb1450 [DOI] [PubMed] [Google Scholar]
- 39.Bellisarii FL, Gallina S, De Caterina R. Tumor necrosis factor-alpha and cardiovascular diseases. Ital Heart J. 2001;2(6):408-417. [PubMed] [Google Scholar]
- 40.Liu Y, Zou J, Li B, et al. RUNX3 modulates hypoxia-induced endothelial-to-mesenchymal transition of human cardiac microvascular endothelial cells. Int J Mol Med. 2017;40(1):65-74. doi: 10.3892/ijmm.2017.2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y, Chang AC, Fournier M, Chang L, Niessen K, Karsan A. RUNX3 maintains the mesenchymal phenotype after termination of the Notch signal. J Biol Chem. 2011;286(13):11803-11813. doi: 10.1074/jbc.M111.222331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di W, Lv J, Jiang S, et al. PGC-1: the energetic regulator in cardiac metabolism. Curr Issues Mol Biol. 2018;28:29-46. doi: 10.21775/cimb.028.029 [DOI] [PubMed] [Google Scholar]
- 43.Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30(4):145-151. doi: 10.1152/advan.00052.2006 [DOI] [PubMed] [Google Scholar]
- 44.Chen P, Piaggi P, Traurig M, et al. Differential methylation of genes in individuals exposed to maternal diabetes in utero. Diabetologia. 2017;60(4):645-655. doi: 10.1007/s00125-016-4203-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489-499. doi: 10.1038/s41586-019-1411-0 [DOI] [PubMed] [Google Scholar]
- 46.Domingo-Relloso A, Riffo-Campos AL, Haack K, et al. Cadmium, smoking, and human blood DNA methylation profiles in adults from the Strong Heart Study. Environ Health Perspect. 2020;128(6):67005. doi: 10.1289/EHP6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crocker KC, Domingo-Relloso A, Haack K, et al. DNA methylation and adiposity phenotypes: an epigenome-wide association study among adults in the Strong Heart Study. Int J Obes (Lond). 2020;44(11):2313-2322. doi: 10.1038/s41366-020-0646-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langsted A, Bojesen SE, Stroes ESG, Nordestgaard BG. AHRR hypomethylation as an epigenetic marker of smoking history predicts risk of myocardial infarction in former smokers. Atherosclerosis. 2020;312:8-15. doi: 10.1016/j.atherosclerosis.2020.08.034 [DOI] [PubMed] [Google Scholar]
- 49.Philibert RA, Dogan MV, Mills JA, Long JD. AHRR methylation is a significant predictor of mortality risk in Framingham Heart Study. J Insur Med. 2019;48(1):79-89. doi: 10.17849/insm-48-1-1-11.1 [DOI] [PubMed] [Google Scholar]
- 50.Reynolds LM, Wan M, Ding J, et al. DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet. 2015;8(5):707-716. doi: 10.1161/CIRCGENETICS.115.001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Schöttker B, Florath I, et al. Smoking-associated DNA methylation biomarkers and their predictive value for all-cause and cardiovascular mortality. Environ Health Perspect. 2016;124(1):67-74. doi: 10.1289/ehp.1409020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klerk M, Verhoef P, Clarke R, Blom HJ, Kok FJ, Schouten EG; MTHFR Studies Collaboration Group . MTHFR 677C→T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA. 2002;288(16):2023-2031. doi: 10.1001/jama.288.16.2023 [DOI] [PubMed] [Google Scholar]
- 53.Bomsztyk K, Denisenko O, Wang Y. DNA methylation yields epigenetic clues into the diabetic nephropathy of Pima Indians. Kidney Int. 2018;93(6):1272-1275. doi: 10.1016/j.kint.2018.02.015 [DOI] [PubMed] [Google Scholar]
- 54.Hinterseher I, Erdman R, Elmore JR, et al. Novel pathways in the pathobiology of human abdominal aortic aneurysms. Pathobiology. 2013;80(1):1-10. doi: 10.1159/000339303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hattab MW, Clark SL, van den Oord EJCG. Overestimation of the classification accuracy of a biomarker for assessing heavy alcohol use. Mol Psychiatry. 2018;23(11):2114-2115. doi: 10.1038/mp.2017.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eTable 1. Descriptive characteristics for eligible Strong Heart Study participants versus finally selected participants
eTable 2. Strong Heart Study baseline participants’ characteristics by coronary heart disease (CHD) incidence status
eTable 3: Hazard ratios (95% CIs) of CpGs available in the Illumina 450 000 platform initially associated with CHD by elastic-net in the Strong Heart Study (SHS), subsequently associated in the Women’s Health Initiative (WHI), the Framingham Heart Study (FHS) and the Atherosclerosis Risk in Communities (ARIC) Study, and not statistically significant in the meta-analysis
eTable 4. Hazard ratios (95% confidence intervals) of incident coronary heart disease comparing the 90th vs 10th percentile of differentially methylated CpGs available in the Illumina 450 000 platform and selected by untargeted elastic-net in each of the study cohorts, replicated with targeted elastic-net models in at least three of the other cohorts, and statistically significant in the meta-analysis
eTable 5: Targeted approach for the association of differentially methylated CpGs associated with atherosclerotic cardiovascular disease in previous studies and their association with incident CHD in the Strong Heart Study
eTable 6: Untargeted approach (EWAS) for coronary heart disease in the Strong Heart Study and replication in the Framingham Heart Study and the Women’s Health Initiative
eTable 7: Differentially methylated regions (DMRs) associated with incident coronary heart disease (CHD) in the Strong Heart Study
eFigure 1. Flowchart of the data exclusion process of the Strong Heart Study
eFigure 2. Protein-protein interaction network of genes annotated to DMPs selected by untargeted elastic-net models in two or more cohorts
eFigure 3: Hazard ratio (95% confidence intervals) for coronary heart disease and several hypermethylated CpGs annotated to PLEK in the Strong Heart Study
eReferences
eTable. Differentially Methylated Positions (DMPs) selected by an untargeted elastic-net model in the Strong Heart Study using all 450 000 CpGs available


