Figure 11.
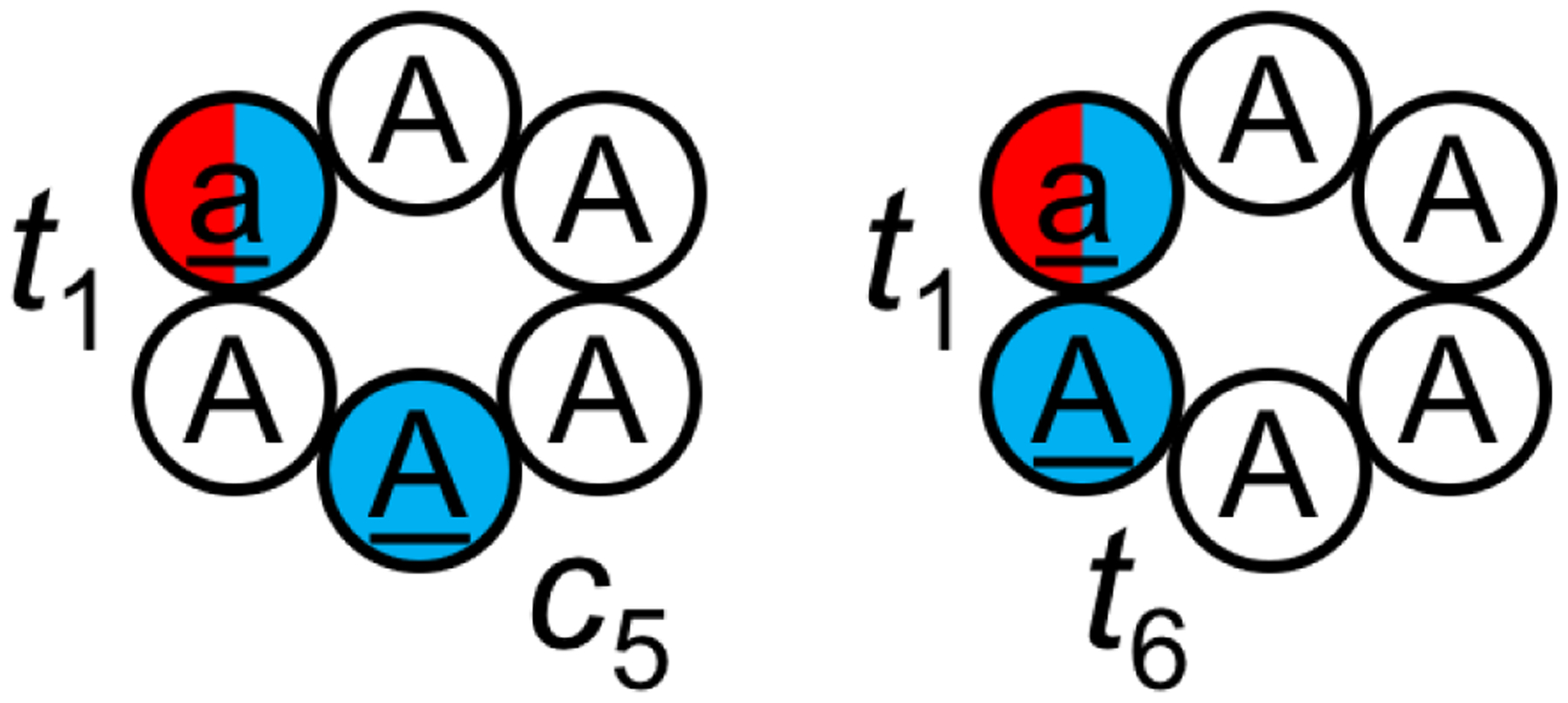
Cyclo-(aAAAAA) and cyclo-(aAAAAA) were simulated using BE-META.239 D-amino acids are listed in lowercase and shown in red; N-methylated amino acids are underlined and shown in blue. In cyclo-(aAAAAA), residues 1 and 5 were N-methylated and the N-methylated amide bonds adopted a trans and cis configuration, respectively (denoted as t1c5). In cyclo-(aAAAAA), residues 1 and 6 were N-methylated and the two N-methylated amide bonds both adopted a trans configuration (denoted as t1t6). It was found that if the correct isomer states were given, the RSFF2 force field was able to reproduce the solution NMR structures for both cyclic peptides; however, when the correct isomer states were not given, the RSFF2 force field was unable to predict the t1c5 isomer of cyclo-(aAAAAA).
