Figure 7. PtdIns4P directly binds SKIP and recruits it to the phagolysosome.
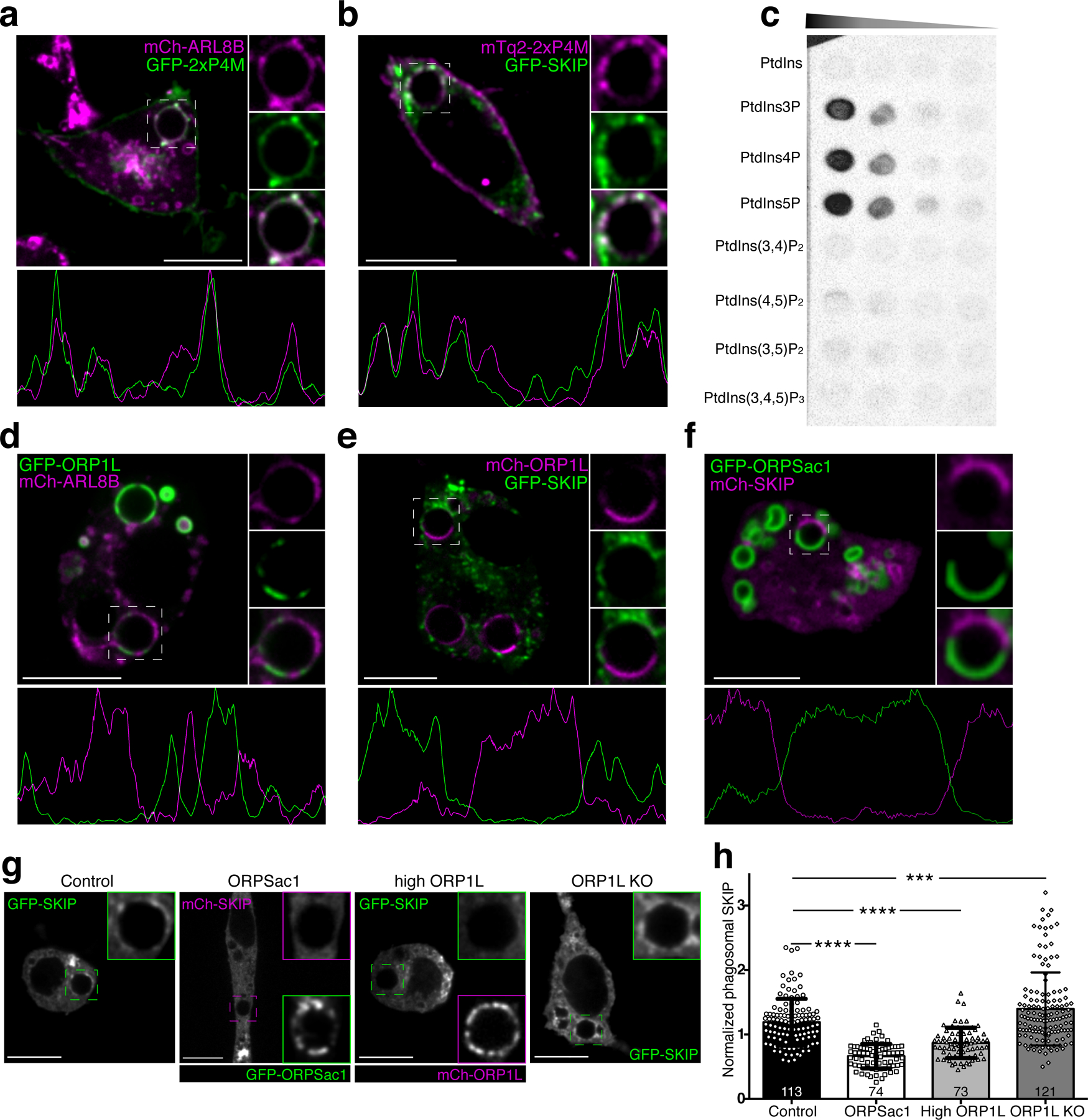
a) Representative confocal micrograph of RAW macrophages co-expressing GFP-2xP4M and mCh-ARL8b after 40 min of phagocytosis; insets show magnifications of the phagosome within the dashed box. The graph below represents a line-scan of fluorescence intensities along the periphery of the phagosome. b) Representative confocal micrograph of RAW macrophages co-expressing mCh-2xP4M and GFP-SKIP after 40 min of phagocytosis; other details as in a. c) Protein overlay assay; a hydrophobic membrane spotted with the indicated lipids was overlaid with purified GST-SKIP (0.3 µg/mL); bound protein was detected by immunoblotting. d) Representative confocal micrograph of RAW macrophages co-expressing GFP-ORP1L and mCh-ARL8b after 40 min of phagocytosis; other details as in a. e) Representative confocal micrograph of RAW macrophages co-expressing mCh-ORP1L and GFP-SKIP after 40 min of phagocytosis; other details as in a. f) Representative confocal micrograph of RAW macrophages co-expressing GFP-ORPSac1 and mCh-SKIP after 40 min of phagocytosis; other details as in a. g) RAW macrophages expressing FP-SKIP after 45 min of phagocytosis of IgG-SRBC: (from left to right) control conditions; expression of GFP-ORPSac1; overexpression of mCh-ORP1L; ORP1L KO. Scale bars = 10 µm throughout. g) Quantitation of SKIP recruitment to phagosomes under the conditions shown in f. Means ± SEM of normalized fluorescent intensities of GFP-SKIP in the number of phagosomes indicated in each bar, from 3 independent experiments. Significance calculated by two-tailed unpaired t tests.
