Fig. 2.
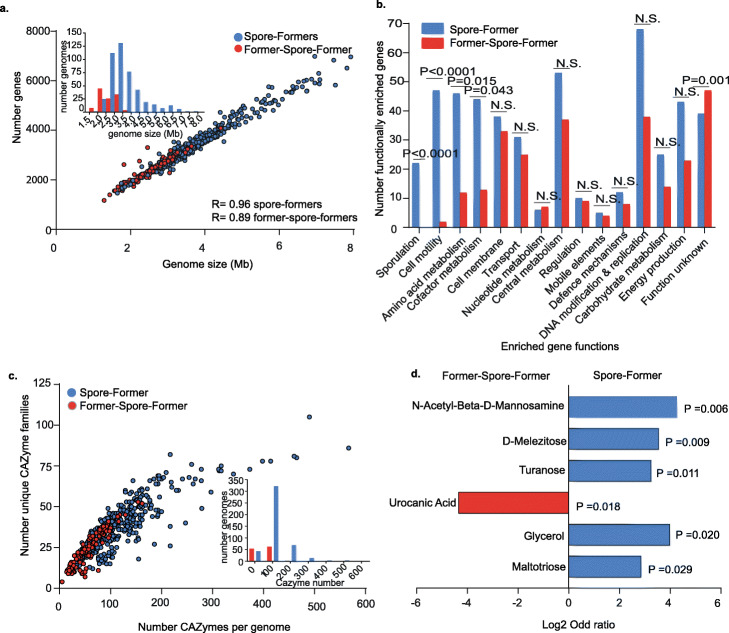
Genome reduction and metabolic specialization during host-adaptation by gut Former-Spore-Formers. a A marker of host adaptation, genomes of gut FSF are smaller than SF genomes (P< 0.0001, Mann-Whitney test), with a strong correlation between genome size and gene number, Spearman rho, R= 0.96 for SF and R= 0.89 for FSF. Inset shows distribution by genome size. b Functional enrichment analysis revealed 489 enriched gut SF genes and 272 enriched FSF genes. Enriched genes were grouped by functional classes. The graph presents the comparison of enriched gene numbers in their functional class and ordered by decreasing statistical significance. Motility, amino acid and cofactor metabolism and sporulation functional classes are statistically more enriched in gut Spore-Formers (SF) compared to Former-Spore-Formers (FSF). No functional classes are more enriched in FSF. Fisher’s exact test, N.S. = not significant. c FSF encode a smaller number of total CAZymes and a smaller number of CAZyme families per genome compared to gut SF (P<0.0001 for both total number and family number, Welch’s t-test). Inset shows the distribution by CAZyme number. d Erysipelotrichaceae FSF have a more restricted carbohydrate utilization profile compared to Erysipelotrichaceae SF. The ability of FSF (n=4) and SF (n=4) to use 95 different carbon sources was tested. N-Acetyl-Beta-D-Mannosamine (P=0.006) (a precursor of sialic acid), D-Melezitose (P=0.009), turanose (P=0.011), glycerol (P=0.020) and maltotriose (P=0.029) are metabolized to a statistically significant greater degree by SF whereas urocanic acid (a derivative of histidine) was metabolized to a statistically significant greater degree by FSF (P=0.018), based on Fisher’s exact test