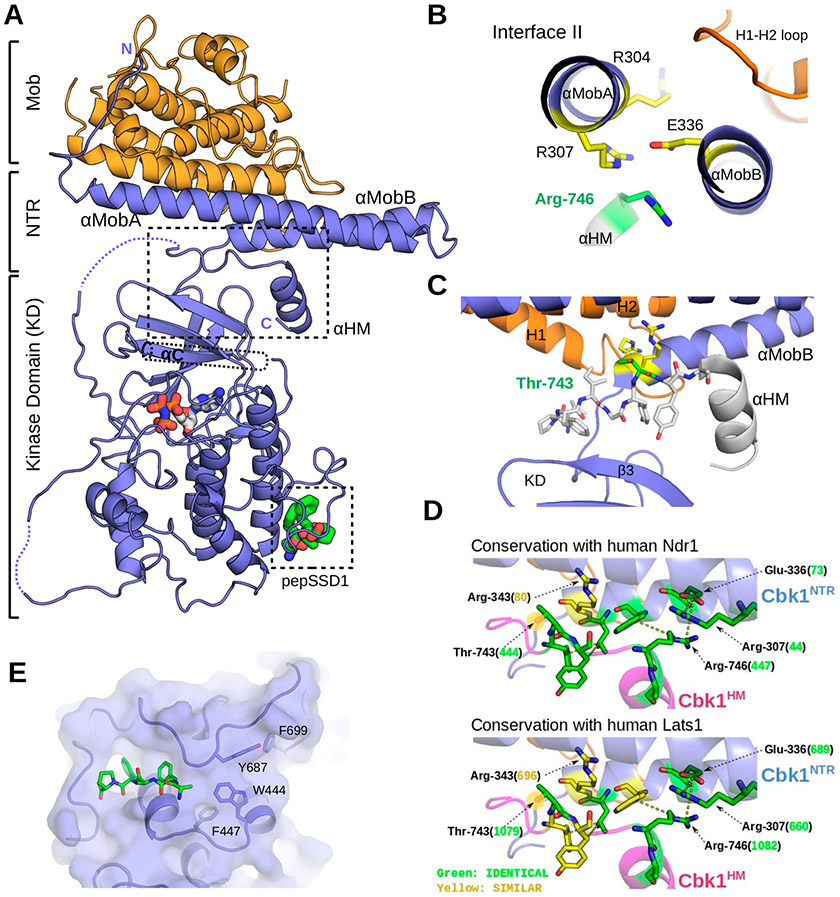
Figure 5.
Revised Cbk1–Mob2 crystal structure that reveals Ndr structural components conserved in Ndr1/Lats1. (A) The crystal structure of the inactive Cbk1–Mob2 complex was remodeled on the basis of the new Cbk1NTR–Mob2 structure. In the new model, the coactivator binds to two N-terminal helices (αMobA and αMobB) that together with the top of the kinase domain (β3) form a binding slot for the AGC kinase hydrophobic motif (HM). The C-terminal part of this motif also adopts a helical conformation (αHM). (B) Cbk1’s hydrophobic motif (HM) binds residues in interface II of the Cbk1NTR. The panel shows the view along the axis of αMobA/B and highlights Arg-746 and critical residues at Cbk1NTR–Mob interface II (Arg-307, Arg-304, and Glu-336). (C) The phosphorylation site on the HM (Thr-743, green) is next to Arg-343 and Arg-344 (yellow) from αMobB, hinting about the importance of Mob coactivator binding in the allosteric phosphoregulation of the Ndr/Lats kinase domain. The HM is shown in stick representation (737-LPFIGYTY-744) or as an α-helical cartoon (745-SRFDYLTKRKNAL-756) in gray. (D) Residues in the interacting region between the HM and the Cbk1NTR are conserved in human Ndr1/Lats1. Green residues are identical; yellow residues are chemically similar. Numbers in parentheses indicate the human Ndr/Lats residue orthologous to the indicated Cbk1 amino acid (black). (E) Crystallographic model rebuilding also improved the overall quality of the electron density and allowed the Ssd1 docking peptide to be traced (green sticks).