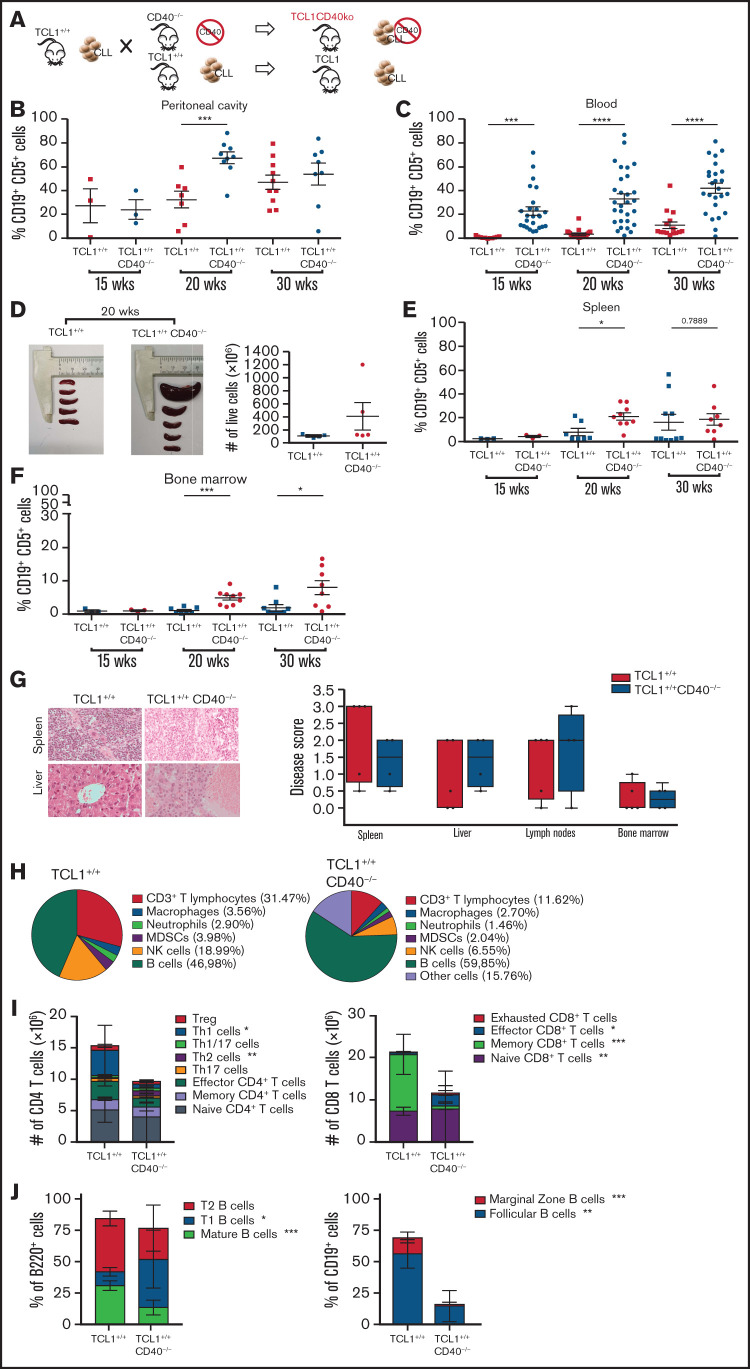
Figure 2.
CD40L is dispensable for CLL induction in the TCL-1 model. (A) Schematic representation of the experiment. TCL-1+/+ and TCL-1+/+CD40−/− mice were analyzed for accumulation of CD19+CD5+ B cells by flow cytometry. Relative contribution (mean ± standard deviation [SD]) of CD19+CD5+ cells to the entire B-cell pool in the peritoneal cavity (B), peripheral blood (C), spleen (E), and bone marrow (F). Each point represents a single mouse. Statistical analysis fo all panels: Student t test, *P < .05, **P < .01, ***P < .001, ****P < .0001. (D) Representative photographs of the spleens (left and middle panels) and cell counts of the whole spleens (right panel) from TCL-1+/+ and TCL-1+/+CD40−/− mice at 20 weeks of age. No statistically significant differences were found. (G) Representative photographs of hematoxylin and eosin–stained tissue sections of the spleen and the liver from TCL-1+/+ and TCL-1+/+CD40−/− mice (magnification ×400; left panel) and CLL disease score (right panel) of hematoxylin and eosin–stained tissue sections from different organs from TCL-1+/+ and TCL-1+/+CD40−/− mice at 20 weeks of age, as defined by a hemopathologist in a blinded fashion based on the degree of leukemia infiltration and disruption of the normal organ architecture in spleen, liver, lymph node, and bone marrow. Boxplots depict the first quartile, median, and third quartile. Vertical bars represent whiskers that indicate the distance from the smallest (lower bar) and the highest (upper bar) nonoutlier values from the first and third quartiles, respectively. Each point represents a single mouse. No statistically significant differences were found using the Student t test. (H) Pie charts show the frequency of the immune infiltrate gated on live cells from the spleen of TCL-1+/+ (left panel) and TCL-1+/+CD40−/− (right panel) mice at 20 weeks of age. Reported cells were identified as follow: CD3+ T lymphocytes (CD3+), macrophages (CD11b+F4/80+), neutrophils (CD11b+Ly6G+), myeloid-derived suppressor cells (MDSCs; CD11b+Gr1+), natural killer (NK) cells (NK1.1+CD3−), and B cells (B220+). All other populations are classified as “Other cells.” (I) Number of CD4 (left panel) and CD8 (right panel) T cells with the described phenotype infiltrating the spleen of TCL-1+/+ and TCL-1+/+CD40−/− mice from panel H. Reported cells were identified as follows: Treg (CD3+CD4+CD25+FOXP3+), Th1 cells (CD3+CD4+CXCR3+CCR4−CCR6−), Th1/17 (CD3+CD4+CXCR3+CCR4−CCR6+), Th2 (CD3+CD4+CXCR3−CCR4+CCR6−), Th17 (CD3+CD4+CXCR3−CCR4+CCR6+), effector CD4+ T cells (CD3+CD4+CD44+CD62L−), memory CD4+ T cells (CD3+CD4+CD44+CD62L+), naive CD4+ T cells (CD3+CD4+CD44−CD62L+), effector CD8+ T cells (CD3+CD8+CD44+CD62L−), memory CD8+ T cells (CD3+CD8+CD44+CD62L+), naive CD8+ T cells (CD3+CD8+CD44−CD62L+), and exhausted CD8+ T cells (CD3+CD8+CD39+PD-1+). Dead cells were excluded by viability dye staining. Bar graphs show mean ± SD (n = 5 mice per group). (J) Frequency of B-cell populations infiltrating the spleen of TCL-1+/+ and TCL-1+/+CD40−/− mice from panel H. Reported cells were identified as follows: mature B cells (B220+IgD+IgMint), T1 B cells (B220+IgD−IgM+), T2 B cells (B220+IgD+IgM+), follicular B cells (CD19+CD23+CD21int), and marginal zone B cells (CD19+CD23−CD21+). Dead cells were excluded by viability dye staining. Bar graphs show mean ± SD (n = 5 mice per group). wks, weeks.