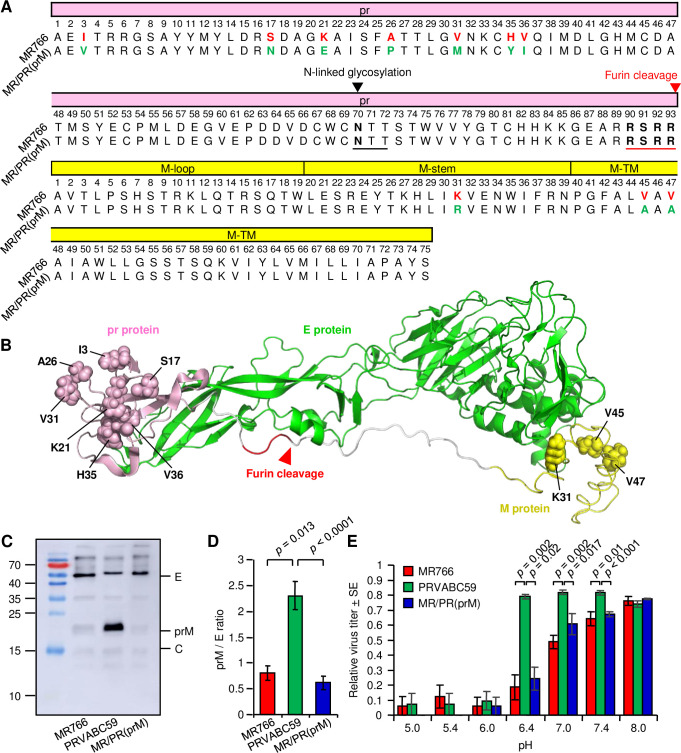
Fig 3. prM cleavage and pH stability.
(A) Sequence alignment of prM from MR766 and MR/PR(prM) (or PRVABC59). The N-linked-glycosylation site (black arrowhead) and motif (underlined) are shown. The furin cleavage site (red arrowhead) and motif (red underline) are indicated. (B) Structure prediction of MR766 prME monomer in the immature virus. The homology model based on dengue virus structure (PDB accession code: 4B03) was generated by MOE homology modeler. The pr, M and E proteins are shown in light pink, yellow and green, respectively. Different amino acid residues in prM protein between MR766 and MR/PR(prM) are shown as atomic spheres. The furin cleavage site (red arrowhead) and motif (red) are indicated. (C) Representative example of a Western blot of purified MR766, PRVABC59 and MR/PR(prM). (D) Densitometry analyses of prM and E proteins from Western blots of purified MR766, PRVABC59 and MR/PR(prM). The intensities of the prM and E bands were determined using ImageJ software, with the mean (7 purified preparations of MR766, 6 purified preparations of PRVABC59 and MR/PR(prM)) prM/E ratio shown. Statistical analysis was performed by t-test. (E) pH stability of MR766, PRVABC59 and MR/PR(prM). MR766 (n = 6 replicates), PRVABC59 (n = 8) and MR/PR(prM) (n = 6) were treated at the indicated pH for 30 min, and virus titers were determined by CCID50 assays. Kolmogorov-Smirnov test or t-test was used for statistical analysis.