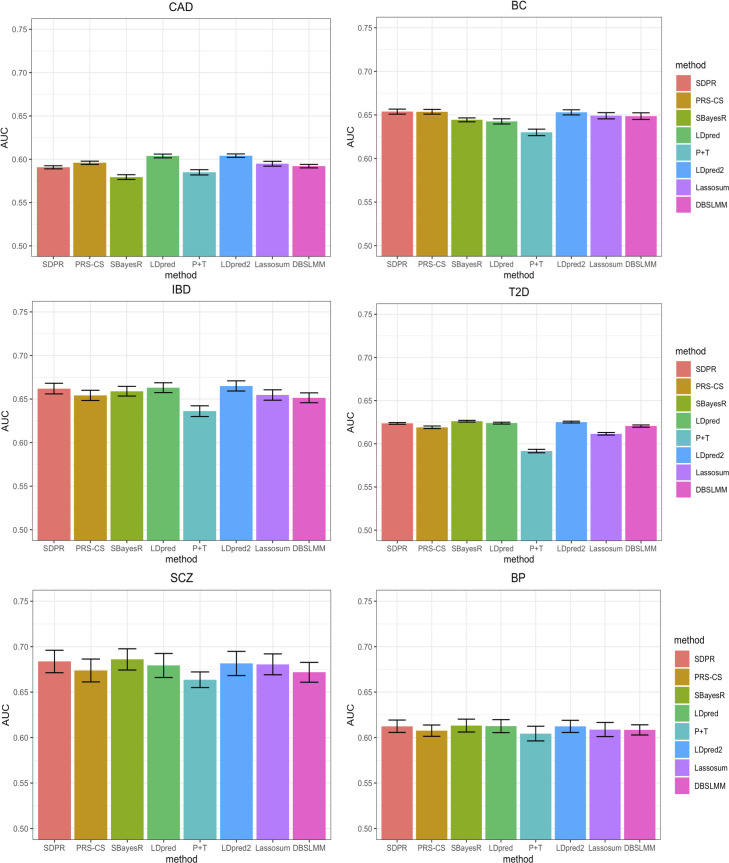
Fig 3. Prediction performance of different methods for 6 diseases in the UK biobank.
Selected participants with corresponding diseases were randomly assigned to form validation and test dataset (Table 1). For PRS-CS, LDpred, P+T, LDpred2, lassosum and DBSLMM, parameters were tuned based on the performance on the validation dataset. We repeated the split and tuning process for 10 times. The mean AUC across 10 random splits was reported in the Table H in S1 Text.