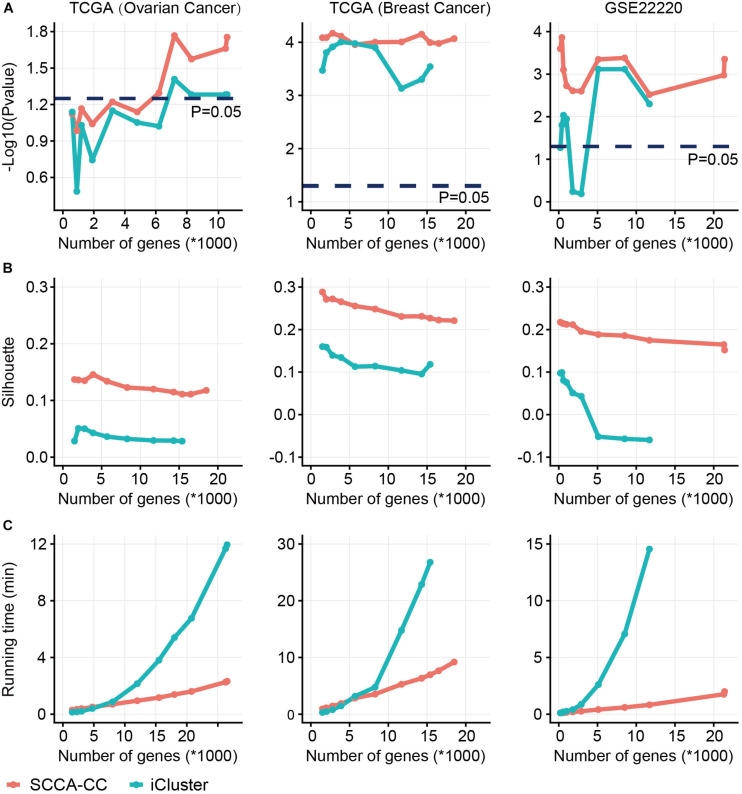
FIGURE 4.
A comparison of sparse canonical correlation analysis for cancer classification (SCCA-CC) with iCluster. Using varying numbers of genes preselected based on MAD, we compared the classification performance between SCCA-CC and iCluster based on (A) P- values indicative of association with survival calculated by log–rank tests, (B) Silhouette score representing the coherence of clusters, and (C) the algorithm running time evaluating the computational complexity.