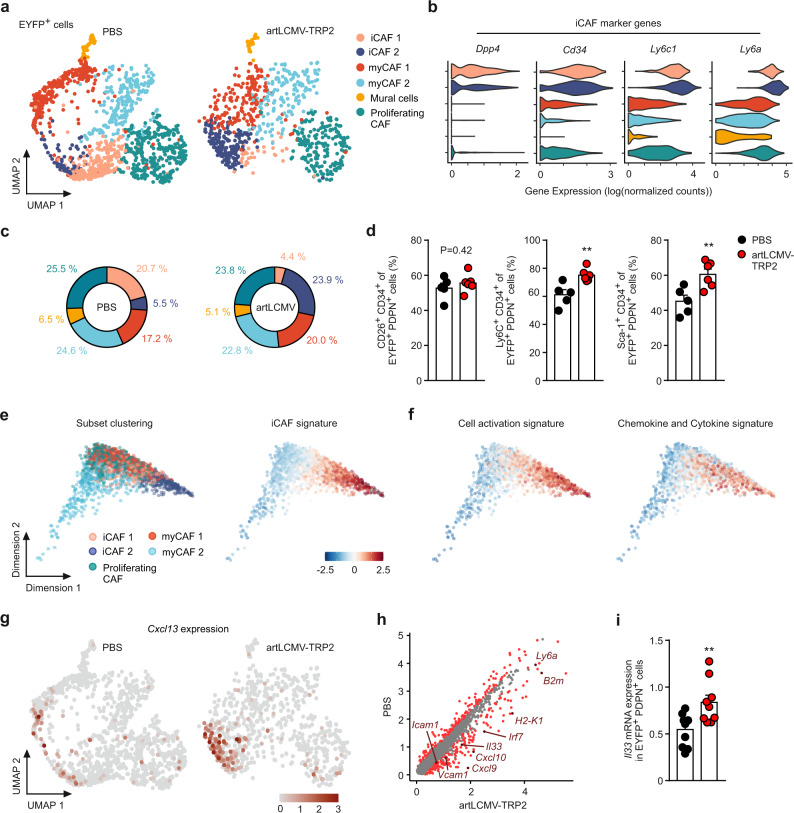
Fig. 5. Activation of Cxcl13-expressing immune-stimulatory FSCs by intratumoral artLCMV-TRP2 treatment.
Single-cell RNA-seq analysis of EYFP+ cells isolated from the tumors of Cxcl13-Cre/ tdTom EYFP mice treated i.t. with artLCMV-TRP2 or PBS on day 11. a UMAP plots displaying FSCs cluster assignment in PBS control or artLCMV-treated mice. b Violin plots depicting expression of iCAF marker genes Ly6c1, Ly6a, Cd34, and Dpp4. c Pie charts showing the relative abundance of the identified FSC clusters. d Flow cytometric analysis of EYFP+ PDPN+ cells stained for CD26 (Dpp4), CD34, Ly6C (Ly6c1), and Sca-1 (Ly6a/ Ly6e). e, f Diffusion maps of EYFP+ tumor FSCs with trajectories constructed based on differential genes analysis in clusters iCAF1 and iCAF2 between PBS and artLCMV condition. e Diffusion maps showing subset clustering and iCAF signatures and f cell activation signature and chemokine and cytokine signature. g UMAP feature plots of Cxcl13 expression. h Scatter plot depicting DE genes of iCAF1 and iCAF2 between PBS control and artLCMV-TRP2-treated mice. i Real-time PCR analysis for Il33 expression in EYFP+ PDPNhi sorted cells from tumors of Cxcl13-Cre/tdTom EYFP mice. ScRNA-seq analysis was performed with one biological replicate for n = 6 PBS-treated mice and two biological replicates for n = 3–4 artLCMV-TRP2-treated mice. We obtained 1293 (PBS) and 786 (artLCMV-TRP2) EYFP-expressing cells. Dots in d, i represent individual mice and shown is mean + s.e.m. values. Pooled data from two independent experiments with n = 5 (PBS) and n = 6 (artLCMV-TRP2) mice (d); n = 10 (PBS) and n = 9 (artLCMV-TRP2) mice (i). Statistical analysis was performed using unpaired two-tailed Student’s t test (d, i) with *P < 0.05: **P < 0.01; ***P < 0.001. Source data and exact P values are provided in the Source data file.