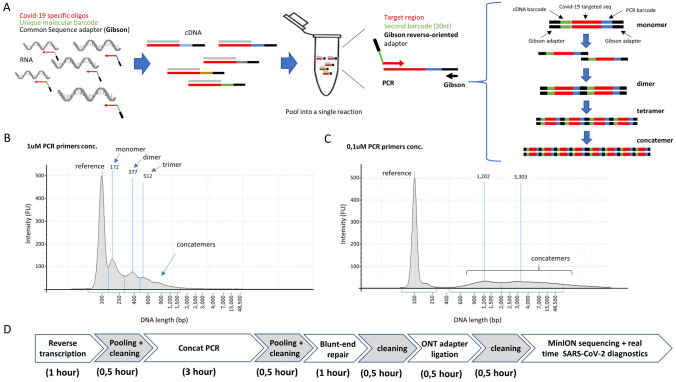
Figure 2.
Dual molecular DNA barcoding and PCR-driven concatemerization applied to SARS-CoV-2 diagnostics. (A) Reverse transcription is performed with oligonucleotides targeting a Covid-19 region, but in addition containing a unique molecular barcode per sample and a common adapter sequence (Gibson). Reverse transcription assays (cDNA) performed for multiple candidates are pooled together for amplification. PCR amplification is performed with oligonucleotides targeting the common Gibson sequence and a Covid-19-specific primer hosting a second molecular barcode as well as a complementary Gibson sequence. During PCR cycles, amplified products are able to concatenate thanks to the Gibson flanking sequences. We have called this particular PCR strategy “Concat-PCR”. (B) DNA electropherogram (Agilent TapeStation system) illustrating concatemer products obtained with PCR primers at 1uM final concentration. (C) DNA electropherogram for a concat-PCR assay performed with PCR primers at 0.1uM final concentration, demonstrating a preferential production of long concatemers in contrast to those observed in (B). (D) Timeline illustrating the various molecular biology steps required for samples preparation prior engaging samples into MinION DNA sequencing.