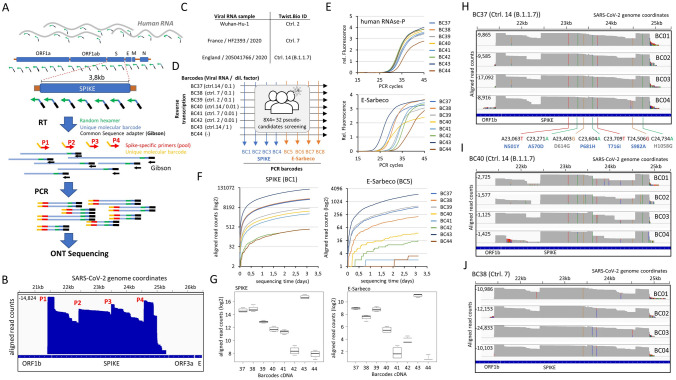
Figure 4.
SARS-CoV-2 variants tracking within the SPIKE gene. (A) Scheme illustrating the SPIKE targeted strategy for variants tracking with Oxford Nanopore technology (ONT) sequencing. Reverse transcription is performed with random hexamer primers presenting unique molecular barcodes and the Gibson adapter sequence. This approach, allows to generate cDNA products of variable length covering the whole viral but also the human genome. Barcoded cDNA samples are PCR amplified with four SPIKE-specific primers (each of them including a common molecular DNA barcode and spaced by ~ 1 kb distance) and one Gibson primer targeting the adapter sequence introduced during reverse transcription. For facilitating the assay, all four SPIKE and Gibson primers are used within an equimolar pool. PCR amplified samples are analyzed by Oxford Nanopore MinION DNA sequencing. (B) Genome browser view displaying the aligned read counts obtained after ONT sequencing. Location of each of the SPIKE-targeting primers (P1-P4) is displayed and corresponds to regions presenting the highest read counts within the SPIKE gene. (C) Summary of the used synthetic SARS-CoV-2 viral RNA samples, corresponding to the Wuhan-Hu-1 control sample (reference strain) and two described variant strains (HF2393 and B.1.1.7). (D) Reverse transcription assay scheme for 7 conditions corresponding to different dilutions of viral RNA issued from the strains illustrated in (C) (BC37-BC43) and a control sample devoid of viral RNA (BC44). In all cases, 15 ng of human RNA material—issued from cell lines in culture—has been also introduced. All 8 samples are pooled at the end of the reverse transcription assay and PCR amplified with either four pools of barcoded primers targeting the SPIKE gene (BC1–BC4) or four barcoded primers targeting the E-Sarbeco region (BC5–BC8), providing a combinatorial barcoding strategy corresponding to 32 pseudo-candidates. (E) Quantitative PCR targeting either the human RNAse-P gene or the viral E-Sarbeco region, performed over the reverse transcription assays described in (D). (F) Real-time Covid-19 diagnostic performed by RETIVAD displaying the number of aligned read counts per barcoded cDNA sample (described in (D)) for the gene SPIKE (BC1) or the E-Sarbeco region (BC5). (G) Boxplots illustrating the total aligned read counts associated to the gene SPIKE or the E-Sarbeco region for each of the conditions described in (D). (H) Variant analysis performed by RETIVAD over the barcoded sample 37 (BC37) associated to the corresponding PCR barcodes (BC1–BC4). The displayed genome browser view reveals the presence of multiple nucleotide variants (colored bars), among them seven being retrieved on all four samples (BC1-BC4) and with a significant read coverage. Five of them (A23,063T; C23,271A; C23,604A; C23,709T; T24,506G) correspond to missense mutations that were described within the B.1.1.7 UK variant strain (blue). In addition, two other missense mutations were also detected; the D614G (previously described within SARS-CoV-2 European strains) and the H1058G. (I) Similar to (H) but for the sample BC40, corresponding to a tenfold lower RNA titer than that used for sample BC37. Note that the same missense mutations retrieved in (H) are observed (colored bars), and this in despite of the lower aligned read counts. (J) Variants detection assay over the sampled BC38, corresponding to the SARS-CoV-2 variant strain HF2393, described in France in 2020. In this case, only the missense mutation D614G is retrieved among all four samples (BC1–BC4).