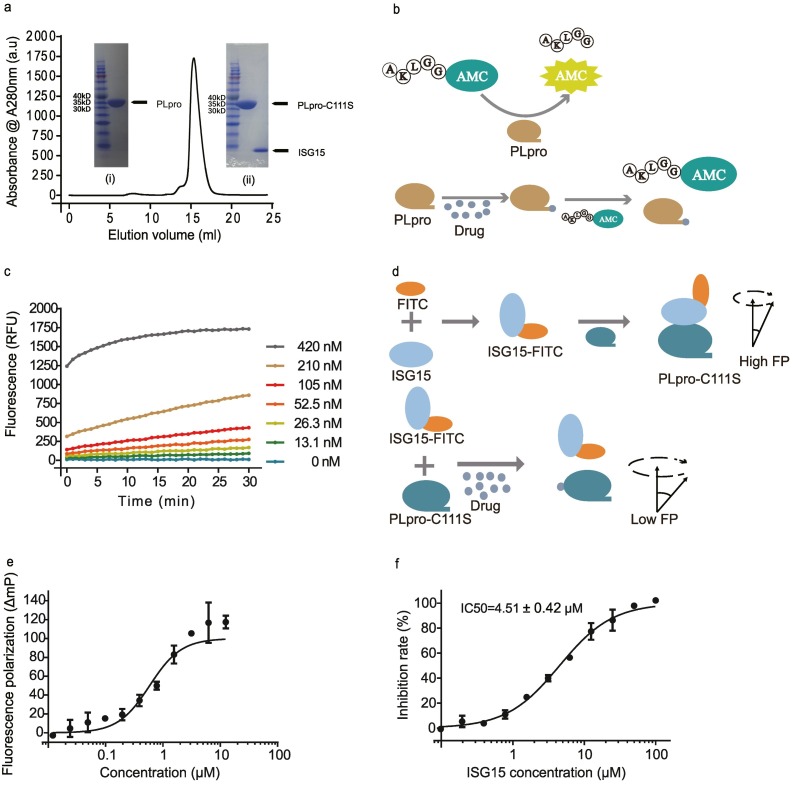
Fig. 1.
Two assays established for screening inhibitors targeting SARS-CoV-2 PLpro. a) Size exclusion chromatography curve of PLpro purification. In the inset (i) the PLpro protein band is present at ~36.8kD on 12% SDS-PAGE, (ii) the PLpro-C111S protein band is present at ~36.8 kD on 12% SDS-PAGE and the ISG15 protein band at ~9.5 kD. b) The schematic diagram for the PLpro protease activity-based assay for inhibitor screening. Cleavage of a fluorogenic peptide by PLpro will release a free AMC fluorophore with its fluorescence signal correlated to the protease cleavage kinetics. The cleavage kinetics will be changed upon inhibition of the protease activity by an inhibitor (i.e., a drug candidate). c) Dependence of reaction kinetics (shown as fluorescence changes) on PLpro concentrations at a constant concentration of ALKGG-AMC (125 μM). d) The schematic diagram for the competitive fluorescence polarization-based assay for inhibitor screening. The binding between inactive C111S mutant of SARS-CoV-2 PLpro with FITC labeled ISG15 will restrict the rotation of ISG15-FITC. This restriction will be indicated with higher fluorescence polarization signals. The competition between an inhibitor and PLpro for binding with ISG15-FITC will be reflected from the concentration dependence of the fluorescence polarization signal. e) Florescence polarization changes in the reaction between PLpro and ISG15-FITC. f) Florescence polarization changes in competition with an unlabeled ISG15.