Abstract
DNA methylation, as an epigenetic mechanism, has a vital role in heart development. An increasing number of studies have investigated aberrant DNA methylation in pediatric or adult heart samples from patients with congenital heart defects (CHD). Placenta tissue, umbilical cord blood, or newborn blood have also been used to detect DNA methylation biomarkers for CHD. However, few studies have compared the methylation levels in fetal heart tissue with cardiac defects with that in normal controls. The present study conducted an integrative whole-genome and CpG site-specific DNA methylation analysis of fetal heart samples from 17 isolated cardiac defect cases, 14 non-isolated cardiac defect cases, and 22 controls with normal hearts, using methylated DNA immunoprecipitation microarray and MassARRAY EpiTYPER assays. Expression of genes adjacent to differentially methylated regions (DMRs) was measured by RT-qPCR and western blot analysis. The results revealed that fetuses with cardiac defects presented global hypomethylation. Genomic analysis of DMRs revealed that a proportion of DMRs were located in exons (12.4%), distal intergenic regions (11.14%), and introns (8.97%). Only 55.7% of DMRs were observed at promoter regions. Functional enrichment analysis for genes adjacent to these DMRs revealed that hypomethylated genes were involved in embryonic heart tube morphogenesis and immune-related regulation functions. Intergenic hypermethylation of EGFR and solute carrier family 19 member 1 (SLC19A1), and intragenic hypomethylation of NOTCH1 were validated in fetal heart tissues with cardiac defects. Only SLC19A1 expression was significantly decreased at the mRNA level, while EGFR, NOTCH1, and SLC19A1 expression were all significantly decreased at the protein level. In conclusion, the present study demonstrated that fetal cardiac defects may be associated with alterations in regional and single CpG site methylation outside of promoter regions, resulting in differentiated expression of corresponding genes associated with heart development. These results present new insights into the epigenetic mechanisms underlying abnormal heart development.
Keywords: fetal, cardiac defects, CpG site methylation, genes expression alterations
Introduction
Heart development is a complex morphological process requiring precise spatiotemporal coordination of diverse cell types. Cardiac defects result from abnormal heart formation and are characterized by various phenotypes, including atrial septum, ventricular septum, and vessel and valve defects. Some categories of severe cardiac defects can cause perinatal morbidity and mortality with a prevalence of 2.5-3 per 1,000 live births (1). Numerous studies have investigated the etiology of cardiac defects. Mutations and differential expression of genes have been identified in several cardiac defect phenotypes (2). Chromosomal and structural DNA abnormalities could be the cause of certain cardiac defects (2). Some environmental risk factors have also been linked to cardiac defects (3). However, for the majority of cases, the etiology has not been fully explored. Extensive studies support the role of the epigenetic mechanisms, as an interplay of genetic and environmental factors in cardiac defects. Therefore, the present study focused on the epigenetic process of cardiac defect formation.
Accumulating evidence suggests that abnormal epigenetic regulation could have an important role in the development of cardiac defects (4). DNA methylation, defined as methylation of cytosine residues at CpG dinucleotides, is one of the most important epigenetic modifications and is involved in a variety of biological process. DNA methylation patterns are continuously changing during early mammalian development, from germlines through to the postnatal stage. Disturbance of these methylation patterns could lead to abnormal development and diseases, including congenital syndromes of immunodeficiency, growth phenotypes, neurodegeneration, and cancer (5). Recently, DNA methylation abnormalities were shown to be a biomarker for the prediction of congenital heart disease and have been explored in neonatal blood spots (6) and placental tissues (7,8). Additionally, in heart tissue samples from patients 1-48 months in age, decreased whole-genome methylation levels, implied by analyzing long interspersed nucleotide element (LINE)1 methylation status, was associated with increased risk of tetralogy of fallot (TOF) (9). Changes in DNA methylation at the promoter and intron regions of several heart development-related genes, including NK2 homeobox 5, GATA binding protein 4, heart and neural crest derivatives expressed 1, and SWI/SNF-related matrix associated actin-dependent regulator of chromatin subfamily a member 4, and their association with the expression of these genes, have also been reported in the myocardium of children with ventricular septal defect (VSD), TOF, and double-chambered right ventricle (10,11). However, limited studies have examined DNA methylation during the fetal period, a key stage when cardiac structures are forming. Furthermore, most current studies have focused on a few simple forms of cardiac defects that are not life-threatening after birth. Such studies are insufficient to provide cues for more severe and complex fetal cardiac defects with or without extracardiac malformations in prenatal life.
The present study was designed to explore the genome-wide DNA methylation landscapes in fetal heart tissues with isolated and non-isolated cardiac defects to identify and validate the global DNA methylation status and CpG site-specific DNA methylation changes adjacent to genes associated with heart development. The expression levels of these heart development-related genes were also evaluated at the mRNA and protein levels.
Materials and methods
Tissue samples
Fetal myocardium tissue was obtained from fetuses whose mothers underwent abortion surgery due to fetal cardiac defects with and without the accompanying noncardiac structure defects (non-isolated and isolated, respectively), diagnosed with fetal echocardiography at Fudan University Affiliated Obstetrics and Gynecology Hospital (Shanghai, China). Normal fetal myocardium tissue samples, as controls, were obtained from fetuses without obvious anatomical abnormalities aborted due to severe maternal complication or trauma. Fetal ventricle myocardium tissue was carefully dissected at the outflow tract area of the fetal heart. Fetal age was calculated based on last menstruation. Gestational age was matched between samples with cardiac defects and controls. The tissue was kept in RNAlater® (Ambion; Thermo Fisher Scientific, Inc.) for at least 24 h at room temperature, then frozen at -80˚C until further use. Thirty-one fetuses with cardiac defects, including 17 isolated cardiac defects and 14 non-isolated cardiac defects, ranging in age from 23 to 27 weeks (mean, 24.5 weeks) and 22 controls with no evidence of cardiac defects ranging in age from 22 to 27.2 weeks (mean, 23.5 weeks) were recruited between January 2011 and December 2012. The sample characteristics and the cardiac defect phenotypes are listed in Table SI, Table SII, Table SIII and Table SIV.
All parents signed written informed consent prior to tissue harvesting for scientific research. The present study was approved by the Ethics Committee of Fudan University Affiliated Obstetrics and Gynecology Hospital (Shanghai, China).
DNA extraction and sodium bisulfite conversion
Genomic DNA was extracted from heart tissue using the QIAamp DNA mini kit (Qiagen GmbH) following the manufacturer's instructions. The DNA concentration and purity were determined by absorbance of A260/A280≥1.8 and A260/A230≥1.9, respectively, using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.). Sodium bisulfite treatment of extracted DNA was performed using an EZ DNA Methylation kit, strictly according to the manufacturer's instructions (Zymo Research, Corp.). The sodium bisulfite-converted DNA was resuspended in 10 µl elution buffer and stored at -20˚C until required for experiments.
Methylated DNA immunoprecipitation microarray (MeDIP-chip) processing and human CpG island microarray
MeDIP-chip analysis was performed using the Roche NimbleGen's DNA methylation assay at CapitalBio Technology, Inc. In brief, 6 µg genomic DNA was digested into 200-1,000 bp fragments with MseI restriction endonuclease. Some of the digested DNA fragments were stored at -20˚C for use as control (input DNA). Some of the remaining digested DNA fragments were immunoprecipitated (IP) with monoclonal anti-5-methyl cytidine antibody (IP DNA), as instructed in the NimbleGen's DNA methylation assay protocol (Roche Diagnostics). Subsequently, IP DNA and input DNA were amplified using the Whole Genome Amplification kit (Sigma-Aldrich; Merck KGaA), following the manufacturer's instructions. Amplified IP and input DNA were labeled with Cy5 and Cy3 dyes, respectively, and co-hybridized to human CGI oligonucleotide microarrays (Roche Diagnostics). The arrays were designed based on the University of California at Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu) CpG island list and contained 27,728 CpG islands plus RefSeq promoters covering 2,440 bp to +610 bp regions from 22,532 potential transcription start sites. Following hybridization and washing, the arrays were scanned using the MS200 scanner (NimbleGen; Roche Diagnostics), then NimbleScan software (version 2.5; NimbleGen) was used to extract the raw fluorescence intensity data from the scanned images. Along with visual inspection, the MS200 scanner and NimbleScan software were applied to show no apparent chip defects and/or that defected areas covered <1% of the array. Further microarray quality assessment was achieved using Sample Tracking Controls features and experimental metrics reports following NimbleGen's instructions on the manufacturer's user guide.
Microarray data analysis
For each probe, after the IP/Input log2 ratio was computed, scaling was performed by subtracting the bi-weight mean from each log2 ratio value on the array. Regions of significant positive enrichment in CHIP-based methylation microarray data were identified using a modified ACME algorithm for peak identification (12). In detail, from the scaled log2 ratio data, a 750 bp fixed-length window was placed around each consecutive probe and the one-side Kolmogorov-Smirnov (KS) test was applied to determine whether the probes had a significantly positive log2 ratio intensity distribution compared with those in the rest of the array. The resulting score for each probe was the -log10 P-value from the windowed KS test around that probe. Regions with more than two probes scoring >2 were defined as MeDIP peaks, and peaks were mapped to genes and CpG islands on the UCSC website (http://genome.ucsc.edu; version NCBI36/hg18).
Bioinformatic analysis
DNA methylation levels of each MeDIP peak CpG region, including several CpG sites, were compared between fetal heart tissues with isolated and/or non-isolated cardiac defects and controls to identify differentially methylated CpG regions (DMRs), using limma, an R package based on linear regression (13). Filtering criteria for statistically significant DMRs were set at P<0.01 and log2 fold change (FC)>0.5. Bedtools (14) was used to identify the LINEs and short-interspersed nuclear elements (SINEs) that overlap with DMRs. Genomic distribution characteristics of these DMRs were analyzed using ChIPpeakAnno workflow in R package (15). Then, DMRs of annotated genes were used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses using gene set information obtained from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/). The R script was from NetBID2 (https://github.com/jyyulab/NetBID). In addition to the mean methylation level across all CpG sites per DMR, the methylation levels of single CpG sites between cardiac defects cases and normal controls were analyzed using the Mann-Whitney nonparametric U test.
Validation of methylation levels by MassARRAY EpiTYPER assay
The Sequenom MassARRAY EpiTYPER platform was used to validate the methylation levels of the targeted regions identified from microarray results (the phenotypes of samples with cardiac defects used for MassARRAY analysis are listed in Table SII). The EpiTYPER assay quantifies CpG dinucleotide methylation based on matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. This is an accurate, sensitive and high-throughput method for the quantitative analysis of DNA methylation at CpG sites (16). The primers used in the present study were designed using Methprimer (http://epidesigner.com; Table SV). Approximately 500 ng of fragmented DNA from each sample was modified by bisulfite treatment. Following PCR with specific primers, which added T7 promoter tags (reverse primer) and 10-mer tags (forward primer), amplicons were treated with shrimp alkaline phosphatase. Fragments were ligated to a T7 promoter segment and transcribed into RNA. The synthesized RNA was cleaved with Rnase A and all cleavage products were analyzed using EpiTYPER software (version 1.0; Sequenom). The detailed protocol was described previously (9).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 100-200 mg of frozen fetal myocardium tissue with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Approximately 500 ng of extracted RNA was used as a template for the reverse transcriptase reaction according to the manufacturers' protocol. Transcribed cDNA was amplified in a qPCR reaction using SYBR Premix Ex Taq system (Takara Biotechnology Co., Ltd.) on the Applied Biosystems 7900 Real-Time PCR system (Thermo Fisher Scientific, Inc.). The amplification reaction conditions were set as follows: An initial 5 min at 94˚C, then 35 cycles comprising 30 sec at 94˚C, 30 sec at 58˚C, and 30 sec at 72˚C, followed by 72˚C for 3 min. Gene expression comparisons were conducted for epidermal growth factor receptor (EGFR), solute carrier family 19 member 1 (SLC19A1), and NOTCH1. All assays were performed in triplicate. GAPDH was used as a housekeeping gene for normalization and the results were analyzed using the 2-ΔΔCq method (17) after averaging the triplicates of each assay. The primers used are listed in Table SVI.
Western blot analysis
Heart tissues from fetuses with and without cardiac defects were homogenized in RIPA lysis buffer (Beyotime Institute of Biotechnology), using an electric homogenizer to break tissue until there was no visible tissue block. Samples were kept on ice for 20 min to fully lyse the tissues. The lysate was ultracentrifuged at 12,000 x g (20 min, 4˚C) to obtain the supernatant for determination of protein concentration using the BCA method. The protein from each sample was treated with 5X loading buffer, and the concentration was then adjusted to 1 µg/µl by supplementing the remaining volume with protein lysate as diluent. Finally, the protein sample was denatured in a 100˚C water bath for 5 min and loaded onto a gel or stored at -80˚C for later use. For each sample, the proteins in 20 µl were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were blocked with 5% non-fat milk powder for 2 h at room temperature in TBS with 0.05% Tween-20 (TBST) and incubated with primary antibodies overnight at 4˚C. Primary antibodies used were rabbit polyclonal EGFR antibody (1:1,000; cat. no. sc-03; Santa Cruz Biotechnology, Inc.), rabbit polyclonal SLC19A1 antibody (1:1,000; cat. no. ab62302; Abcam), rabbit polyclonal NOTCH1 antibody (1:1,000; cat. no. ab27526; Abcam), and rabbit polyclonal GAPDH antibody (1:2,500; cat. no. ab263962 Abcam) as a control. The blots were rinsed in TBST three times and incubated in HRP-conjugated secondary antibody (1:5,000; cat. no. ab6721; Abcam) for 1 h at room temperature. Immunoreactive proteins were developed in ECL Plus and were exposed using the LAS-3000 system (Fujifilm Wako Pure Chemical Corporation). The densitometry of the bands was calculated using ImageJ (version 1.8.0, National Institutes of Health).
Statistical analysis
The methylation levels of single CpG sites between 31 cases of cardiac defects and 22 cases of normal controls were analyzed using the Mann-Whitney nonparametric U test and presented as median with the interquartile ranges. Differences in gene and protein expression levels were assessed by a two-tailed Student's t-test and presented as the means ± standard deviation. All experiments were repeated three times. P<0.05 was considered as statistically significant. All data were analyzed with GraphPad Prims (version 6.0c; GraphPad Software, Inc.).
Results
Sample allocation
The DNA methylation patterns in fetal cardiac tissue of isolated cardiac defects, non-isolated cardiac defects, and control samples were determined using NimbleGen's whole genomic DNA methylation microarray. Overall, 17 sampled from isolated cardiac defects, 14 samples from non-isolated cardiac defects, and 22 samples with normal histology were included in the present study. The methylation analysis was divided into two parts. In the first part, using the methylation microarray, two isolated cardiac defects samples and three non-isolated cardiac defects samples were compared to four normal controls to obtain a list of DMRs (for detailed information, see Tables SI and SII). In the second part, another 15 isolated cardiac defect samples, 11 non-isolated cardiac defect samples and 18 normal controls were used to validate the regional and CpG site-specific methylation level changes in DMRs and expression changes in annotated genes within the DMR (for detailed information, see Tables SIII and SIV).
Global DNA methylation is downregulated in fetal hearts with cardiac defects
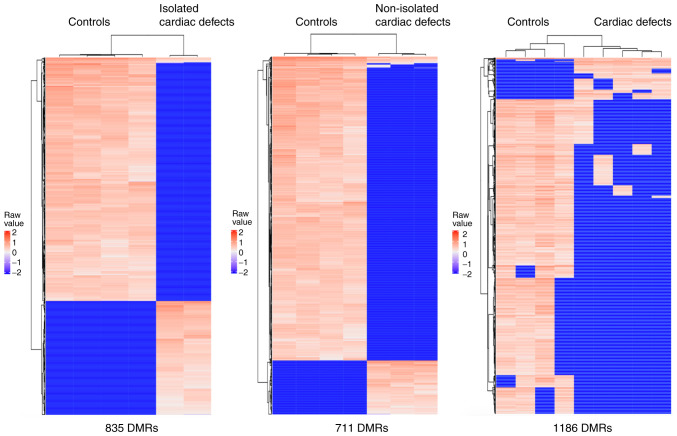
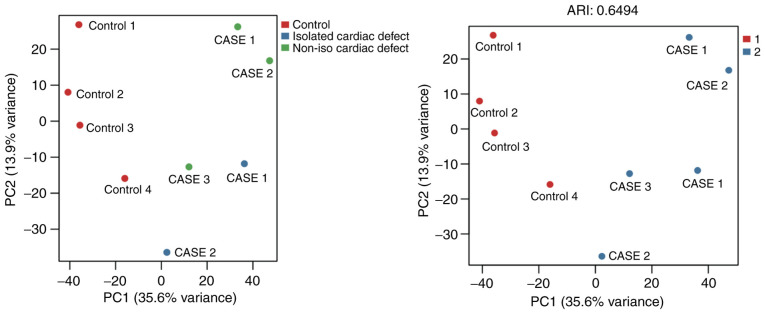
A total of 17,386 CpG sites located in 1,546 methylated regions were captured through the present methylation microarray data analysis. The microarray data have been deposited in the figshare database (DOI: https://doi.org/10.6084/m9.figshare.14130173). The results revealed more hypomethylated regions than hypermethylated regions in both isolated and non-isolated cardiac defect cases compared with the normal controls. In isolated cardiac defect cases, 835 DMRs were tested (Table SVII), of which over 68% (569 DMRs) were hypomethylated (Fig. 1). In non-isolated cardiac defects, 711 DMRs were tested (Table SVIII), of which up to 84% (604 DMRs) were hypomethylated (Fig. 1). After combining isolated and non-isolated cardiac defect cases, the results identified global hypomethylation alteration, at a genome-wide level, with 1,186 DMRs (Table SIX), of which 1,052 (89%) were hypomethylated compared with those in control samples (Fig. 1). Additionally, the present study conducted a principal component analysis based on the most variable levels of 1,737 CpG methylation, which showed that cardiac defect cases were clearly clustered from the normal controls, but the subgroups of cardiac defects with and without extracardiac defects did not appear to differentiate from each other very well (Fig. 2). Therefore, the isolated cardiac defect and the non-isolated cases were combined to form the full set of cardiac defects in subsequent analyses.
Figure 1.
Heatmaps of DMRs showing relative hypomethylation levels in fetal heart with isolated cardiac defects, non-isolated cardiac defects, and cardiac defects irrespective of whether they are isolated. DMRs, differentially methylated regions.
Figure 2.
Principal component analysis for the most variable methylated levels of 1,737 CpG sites. The red dots represent the normal controls. The blue and green dots represent samples with cardiac defects. PC, principal component; ARI, Adjusted Rand index.
To further determine the global alterations of hypomethylation in fetal heart tissue with cardiac defects, the methylation levels of LINEs and SINEs were assessed, because methylation levels of these elements are highly correlated with global DNA methylation (18). In heart tissue from cardiac defect cases, a total of 267 LINEs and 165 SINEs were present in differential methylation profiles, and 75% (200/267) of LINEs and 82% (136/165) of SINEs were hypomethylated.
To validate the reliability of the present DNA methylation alteration patterns of fetal heart tissues with cardiac defects, data from a previous study using the Illumina Infinium HumanMethylation450 BeadChips (450 K arrays) platform, analyzing 49 pediatric human cardiac tissue samples from patients with different CHDs (19), were compared with the MeDIP-chip data from the present study. Of the 1,168 DMRs, two CpG islands, located on chr10:123356616-123358285 and chr13:113772727-113773012, displayed an overlap to the 450 K assay target ID of Hoff et al (19).
Genomic features of DMRs and functional enrichment analysis of genes adjacent to DMRs
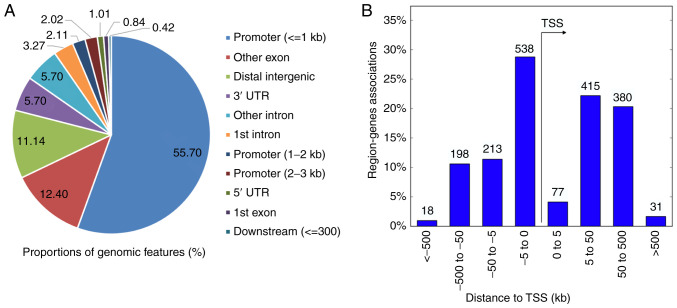
The genomic location of the 1,168 DMRs was examined, the distribution of which is shown in Fig. 3. As the present microarray platform explored CpG islands, the majority (59.83%) of the DMRs were observed in promoters. A considerable number of DMRs were associated with exons (13.24%), introns (8.97%), and distal intergenic areas (11.14%). Regions up to 300 bp downstream of transcriptional start sites, the 5' untranslated region (UTR), and the 3'UTR overlapped with 7.13% of DMRs.
Figure 3.
Genomic distribution of 1,186 DMRs across the genome. (A) The pie chart shows the proportions of genomic features (%). (B) The histogram shows the locations of DMRs in relation to the TSS. DMRs, differentially methylated regions; TSS, transcription start site; UTR, untranslated region.
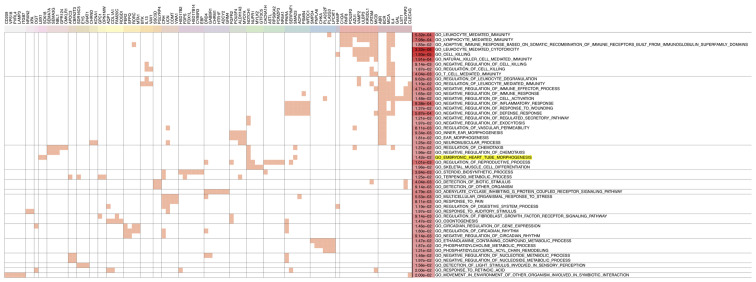
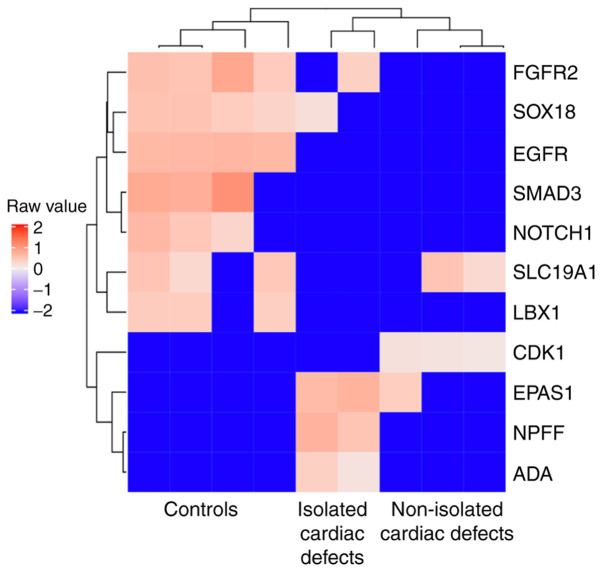
Next, GO analysis was performed for genes contiguous to the 1,168 DMRs to identify significant functional enrichment. The results revealed that most hypomethylated region-related genes were enriched for immune-related functions, suggesting that the immune regulation system may have a role during heart development (Fig. 4). The GO term ‘embryonic heart tube morphogenesis’ was also found to be enriched (Fig. 4). A total of 11 genes, including fibroblast growth factor receptor 2, SRY-box transcription factor 18, epidermal growth factor receptor (EGFR), SMAD3, NOTCH1, solute carrier family 19 member 1 (SLC19A1), ladybird homeobox 1, cyclin-dependent kinase 1, endothelial PAS domain protein 1, neuropeptide FF-amide peptide precursor, and adenosine deaminase, were enriched in GO term ‘embryonic heart tube morphogenesis’ and the associated DMRs were used to generate a corresponding heatmap (Fig. 5). However, no significant heart development-associated enrichment was observed for hypermethylated region-related genes (data not shown).
Figure 4.
Functional enrichment analysis of 1,052 hypomethylated differentially methylated regions related to annotated genes in fetal heart with cardiac defects. Each row represents one GO term. Each column represents one gene. The GO term ‘embryonic heart tube morphogenesis’ is highlighted in yellow. GO, gene ontology.
Figure 5.
Heatmap of 11 differentially methylated regions corresponding to annotated genes which are associated with heart development based on enrichment analysis. FGFR2, fibroblast growth factor receptor 2; SOX18, SRY-box transcription factor 18; EGFR, epidermal growth factor receptor; SLC19A1, solute carrier family 19 member 1; LBX1, ladybird homeobox 1; CDK1, cyclin-dependent kinase 1; EPAS1, endothelial PAS domain protein 1; NPFF, neuropeptide FF-amide peptide precursor; ADA, adenosine deaminase.
Validation of regional and site-specific DNA methylation levels in cardiac defects
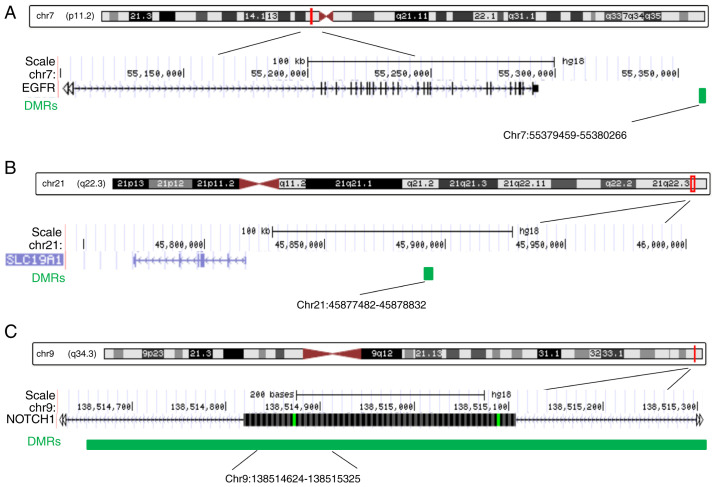
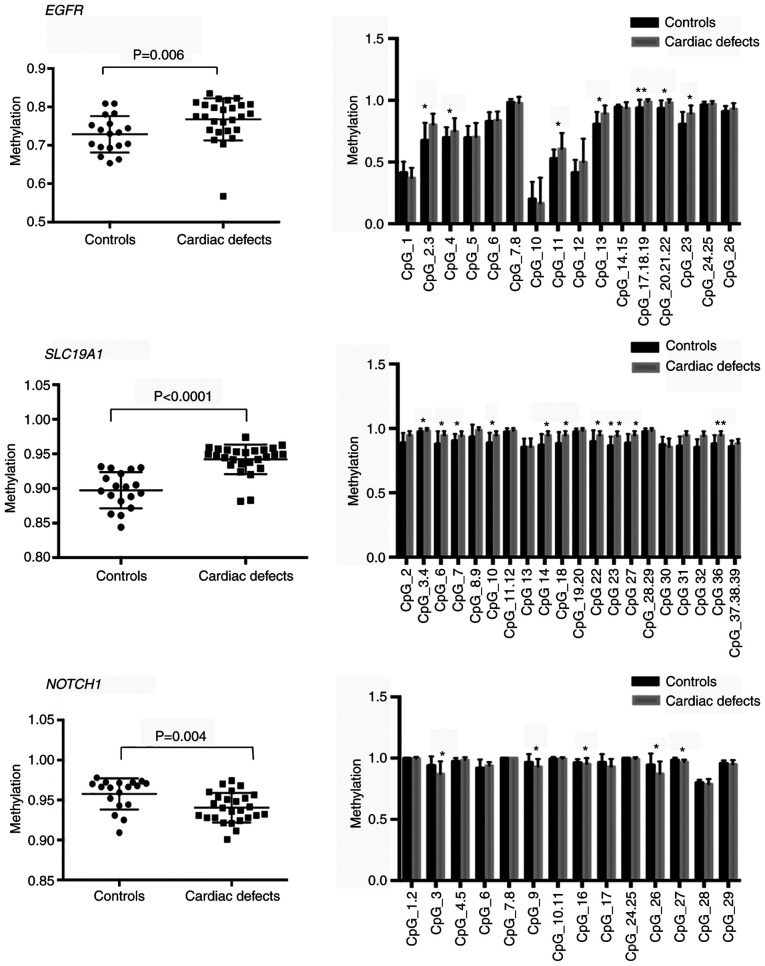
Among the 11 DMRs annotated to heart development-related genes, the three most significant DMRs were selected for further study. Two of the three DMRs, including the EGFR-related and the SLC19A1-related DMRs, were located in intergenic regions, while the NOTCH1-related DMR was intragenic, located in the gene body (Fig. 6). Next, the average methylation levels across all CpG sites of these three DMRs and the methylation levels at single CpG sites were measured. To do this, quantitative methylation analysis was used through the MassARRAY platform in fetal heart tissue samples from an additional 26 cases with cardiac defects, containing 15 isolated cardiac defects and 11 non-isolated cardiac defects, and 18 normal controls. For EGFR-associated DMRs and SLC19A1-associated DMRs, the results demonstrated significantly higher methylation levels in cardiac defect cases compared with those observed in controls [median 0.78 vs. 0.75, interquartile range (IQR) 0.76-0.81 vs. 0.7-0.78, P=0.006 for EGFR DMRs; and median 0.95 vs. 0.91, IQR 0.94-0.96 vs. 0.88-0.93, P<0.0001 for SLC19A1 DMRs) (Fig. 7). The mean CpG region methylation level in the NOTCH1 gene body was significantly lower in cardiac defects cases than in controls (median 0.94 vs. 0.97, IQR 0.93-0.96 vs. 0.94-0.97, P=0.0043; Fig. 7). Specifically, hypermethylation of seven CpG sites, including CpG2.3, CpG4, CpG11, CpG13, CpG17.18.19, CpG20.21.22, and CpG23, at EGFR intergenic regions, and 10 CpG sites, including CpG2, CpG6, CpG7, CpG10, CpG14, CpG18, CpG22, CpG23, CpG27, and CpG36, at SLC19A1 intergenic regions contributed to the increased methylation level in EGFR and SLC19A1 DMRs (Fig. 7). Hypomethylation in the NOTCH1 gene body was mainly attributed to decreased methylation levels of multiple CpGs, including CpG3, CpG9, CpG16, CpG26, and CpG27 (Fig. 7).
Figure 6.
Detailed schematic of CpG sites located in DMRs as summarized by DNA methylation levels in the downstream regions of (A) EGFR and (B) SLC19A1, and (C) in the exonic region of NOTCH1. DMRs, differentially methylated regions; EGFR, epidermal growth factor receptor; SLC19A1, solute carrier family 19 member 1.
Figure 7.
Percentage methylation comparisons between controls (n=18) and cases with cardiac defects (n=26) at differentially methylated regions corresponding to EGFR, SLC19A1, and NOTCH1. Columns represent mean percentage methylation values, and the error bars represent standard deviation. *P<0.05 and **P<0.001. EGFR, epidermal growth factor receptor; SLC19A1, solute carrier family 19 member 1.
EGFR, NOTCH1, and SLC19A1 expression changes at mRNA and protein levels in isolated cardiac defects
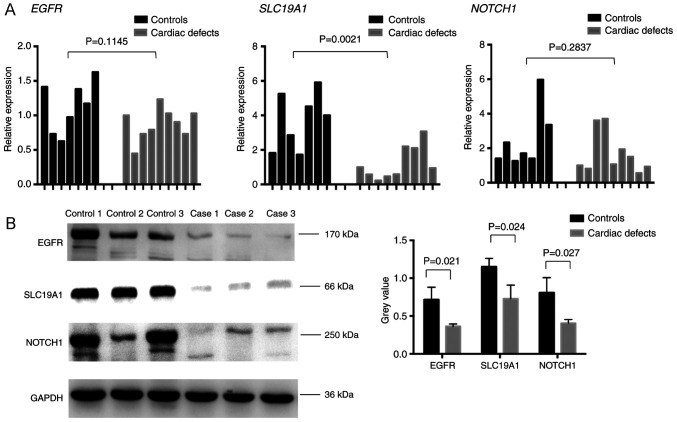
Since changes in global and CpG site-specific methylation levels were observed, the present study analyzed the corresponding gene expression levels. Given the low residual amount of fetal heart tissue, the mRNA expression changes in EGFR, NOTCH1, and SLC19A1 were examined in seven normal controls and nine isolated cardiac defect cases from the previously analyzed sample cohort. Of these, three normal controls and three isolated cardiac defects samples with the most variable mRNA expression levels were used to detect EGFR, NOTCH1, and SLC19A1 protein expression levels. RT-qPCR results showed a statistically significant reduction in SLC19A1 mRNA expression levels in cardiac defects samples compared with normal controls, which was consistent with protein expression results (Fig. 8). Similarly, both EGFR and NOTCH1 displayed a tendency for decreased mRNA expression levels and a significant decline in protein expression in isolated cardiac defects compared with controls (Fig. 8). Notably, changes in the methylation levels observed at EGFR and SLC19A1 intergenic regions showed a trend opposite to that of the expression of the corresponding gene. A consistent trend in the methylation level changes in the NOTCH1 intragenic region and corresponding gene expression levels was identified.
Figure 8.
Validation of EGFR, SLC19A1, and NOTCH1 expression. (A) mRNA expression levels were examined by reverse transcription-quantitative PCR in single fetal hearts from seven normal controls (black bars) and nine cases with cardiac defects (gray bars). (B) Representative images and quantification of protein expression levels as determined by western blot analysis. EGFR, epidermal growth factor receptor; SLC19A1, solute carrier family 19 member 1.
Discussion
Cardiac defects are one of the most prevalent congenital abnormalities of complicated etiology, and their phenotypes vary greatly. Different types of cardiac defects arise from distinct disruptions in heart development at particular stages of heart formation (20). Given that epigenetic regulation is time-specific, it is necessary to understand the underlying mechanisms using samples of the developing heart that are approaching developmental time points when cardiac defects occur. To the best of our knowledge, this genome-wide methylation analysis in heart tissues from a cohort of fetuses is the first of its kind. The present results revealed a lower global methylation status in fetal heart tissue with isolated and with non-isolated cardiac defects compared with normal control tissues. In addition, in heart tissues with fetal cardiac defects, three sets of differentially methylated CpG sites were identified, including DMRs from EGFR and SLC19A1 intergenic regions and a DMR from the NOTCH1 gene body. DNA methylation changes in these DMRs may affect corresponding gene expression and contribute to cardiac defects.
The current study represents a genome-wide methylation analysis of relatively complicated cardiac defects. Previous DNA methylation studies were limited to analyses of methylation status in some congenital heart defect candidate genes (10,11,21-23), and were insufficient to reveal information for DNA methylation on a global scale. One previous study conducted a genome-wide DNA methylation study (24) and found more hypermethylated than hypomethylated DMRs in myocardial samples from pediatric patients with CHDs compared with controls. This is contrary to the present results that revealed a global hypomethylation status in heart tissue with cardiac defects. However, the previous study selected only two CHDs, namely TOF and VSD, obtained heart samples from pediatric patients during surgery, and lacked appropriate age-matched normal hearts. Apart from the above two subtypes of CHD, several complicated and severe CHD phenotypes, possibly exerting a greater impact on pediatric morbidity and mortality, require more in-depth research. In another study (25), whole genome methylation profiles were obtained using heart tissue DNA from fetuses presenting a variety of cardiac defect phenotypes including double outlet right ventricle, hypoplasia of the ascending aorta, right heart hypoplasia, left heart hypoplasia, mitral valve atresia, aorta valve atresia, tricuspid valve stenosis, transposition of the great arteries, and absent ductus arteriosus, but no significant differences in whole genome methylation pattern were identified between fetuses with isolated cardiac defects and fetuses with normal development. Nevertheless, they identified several CpG sites that were differentially methylated in single CHD cases, but these results were not validated in an independent cohort (25).
The global hypomethylation status of fetuses with cardiac defects reported in the present study is consistent with the results of three previously published studies assessing LINE-1 methylation in venous blood samples from very young children with complex CHD (26), mothers whose pregnancies were affected by non-syndromic CHD (27), and in right ventricular tissue samples from pediatric patients (9). The current observation of a global hypomethylation change in cardiac defects is supported by an assessment of increased hypomethylated LINE-1 in fetal heart tissue with cardiac defects and by the detection of a larger proportion of hypomethylated DMRs through whole-genome methylation analysis. Furthermore, detailed comparison of methylation levels in two intergenic DMRs and one intragenic DMR revealed several CpG sites within these DMRs that significantly differed between cases with cardiac defects and controls. The role of CpG site methylation in regions outside of promoters has recently been well-studied. Irrespective of where it is located in the genomic context, DNA methylation serves a crucial role in the transcriptional and splicing regulation of genes (28). To date, several studies have confirmed that intragenic methylated CpG islands are largely tissue-specific and that they are strongly associated with transcription initiation and elongation (28-30). Additionally, discrete hypomethylated regions in intergenic spaces are more predictive of nearby gene activity than are the promoter regions themselves (31). Intergenic DNA hypomethylation, as a downstream target of some signaling pathways, could explain neoplastic tissue overgrowth and developmental disorders (32). Therefore, it is reasonable to speculate that there might be a link between intergenic CpG site methylation levels of EGFR and SLC19A1 DMRs, intragenic CpG sites of NOTCH1 DMRs, and gene expression in the current study. On the other hand, changes in the opposite or same direction between methylation levels of intergenic EGFR and SLC19A1 DMRs and intragenic NOTCH1 DMRs and related gene expression varied based on the genome location and nearby regulatory elements, which was also supported by a previous report (29). This emphasizes the importance of methylation in non-promoter regions. However, the exact mechanisms underlying the intergenic and intragenic methylation-based regulation of EGFR, SLC19A1, and NOTCH1 expression in the present study remain unclear and need to be further explored.
In the present study, RT-qPCR and western blot analyses revealed that EGFR, SLC19A1, and NOTCH1 expression were decreased in fetal heart tissue compared with control normal tissues. These three genes are crucial for heart development. For instance, in mouse studies, Egfr regulates embryonic formation of the aortic valve (33). Slc19a1 knock-out mice were shown to die post-implantation at E6.5, even with folic acid supplementation, and the mice presented with cardiac malformations, including VSDs, thin myocardial wall and VSD with overriding aorta (34). Additionally, Notch signaling is required for cardiac fate determination, patterning of the primitive heart (3), and cardiac valve morphogenesis (35). Mutations in NOTCH1 have been associated with left ventricular outflow tract malformations, including aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome (36). Based on these previous studies, it can be inferred that hypermethylation of EGFR and SLC19A1 intergenic regions and hypomethylation of NOTCH1 intragenic regions might be associated with downregulation of gene expression, resulting in the occurrence of fetal cardiac defects.
Of note, immune regulation was one of the most highly enriched biological functions identified in the present GO enrichment analysis. Previously, the function of an endogenous complement inhibitor, which had an impact on cardiac neural crest cell migration in the zebrafish model, was described, suggesting that immune system molecules may be involved in cardiac tissue development (37). Single-cell transcriptome analysis in the human fetal heart by Cui et al (38) confirmed the role of immune cells in heart development and found that the proportion of these cells greatly increased with the development of the heart.
There are several limitations in the present study. Firstly, the number of cases for some cardiac defect subtypes was small. Secondly, the present study did not use multiple methods to validate the methylation microarray data, because of undetectable methylation level of some CpG sites by EpiTYPER analysis and certain methodological limitations. Thirdly, some interfering factors, including fetal chromosomal abnormalities, the overlap between CpG sites and single-nucleotide polymorphisms and adverse maternal exposure, should also be considered and excluded. Fourthly, subsequent studies are required to clarify the relationship between gene expression and CpG site-specific methylation levels at specific areas and at CpG sites other than those near the EGFR, SLC19A1, and NOTCH1 promoters.
The present study is the first to explore methylation level changes in heart tissue from fetuses with isolated and non-isolated cardiac defects. The results revealed both a global hypomethylation status and absolute changes in DNA methylation at particular CpG sites for cardiac defect samples. The mRNA and protein expression analyses of heart development-associated genes adjacent to DMRs may provide new insights into the possible epigenetic mechanisms of heart development during the fetal period. In the future, it would be worth examining the causality of the three identified DMRs, especially the CpG sites and related gene expression.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by grants from the Shanghai Key Program of Clinical Science and Technology Innovation (grant nos. 17411950500, 17411950501 and 18511105602), the National Natural Science Foundation of China (grant nos. 81901500, 81741047, 81971411 and 81801468), and the Shanghai Medical Center of Key Programs for Female Reproductive Diseases (grant no. 2017ZZ01016).
Availability of data and materials
The datasets generated during the current study are available in the figshare database (https://doi.org/10.6084/m9.figshare.14130173).
Authors' contributions
DM and XL designed the study and provided funding for the study. JZ participated in the experiments and writing. YX participated in sample acquisition and helped to draft the manuscript. XD was responsible for bioinformatics analysis and for generating some of the charts. HW participated in the study concept and design. YQ was responsible for part of the experiments. XL and JZ confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Fudan University Affiliated Obstetrics and Gynecology Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Yuan S, Zaidi S, Brueckner M. Congenital heart disease: Emerging themes linking genetics and development. Curr Opin Genet Dev. 2013;23:352–359. doi: 10.1016/j.gde.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vecoli C, Pulignani S, Foffa I, Andreassi MG. Congenital heart disease: The crossroads of genetics, epigenetics and environment. Curr Genomics. 2014;15:390–399. doi: 10.2174/1389202915666140716175634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarrell DK, Lennon ML, Jacot JG. Epigenetics and mechanobiology in heart development and congenital heart disease. Diseases. 2019;7(52) doi: 10.3390/diseases7030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 6.Bahado-Singh RO, Zaffra R, Albayarak S, Chelliah A, Bolinjkar R, Turkoglu O, Radhakrishna U. Epigenetic markers for newborn congenital heart defect (CHD) J Matern Fetal Neonatal Med. 2016;29:1881–1887. doi: 10.3109/14767058.2015.1069811. [DOI] [PubMed] [Google Scholar]
- 7.Radhakrishna U, Albayrak S, Zafra R, Baraa A, Vishweswaraiah S, Veerappa AM, Mahishi D, Saiyed N, Mishra NK, Guda C, et al. Placental epigenetics for evaluation of fetal congenital heart defects: Ventricular septal defect (VSD) PLoS One. 2019;14(e0200229) doi: 10.1371/journal.pone.0200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahado-Singh R, Vishweswaraiah S, Mishra NK, Guda C, Radhakrishna U. Placental DNA methylation changes for the detection of tetralogy of Fallot. Ultrasound Obstet Gynecol. 2020;55:768–775. doi: 10.1002/uog.20292. [DOI] [PubMed] [Google Scholar]
- 9.Sheng W, Wang H, Ma X, Qian Y, Zhang P, Wu Y, Zheng F, Chen L, Huang G, Ma D. LINE-1 methylation status and its association with tetralogy of fallot in infants. BMC Med Genomics. 2012;5(20) doi: 10.1186/1755-8794-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng W, Qian Y, Wang H, Ma X, Zhang P, Diao L, An Q, Chen L, Ma D, Huang G. DNA methylation status of NKX2-5, GATA4 and HAND1in patients with tetralogy of fallot. BMC Med Genomics. 2013;6(46) doi: 10.1186/1755-8794-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y, Xiao D, Guo X, Chen H, Hao L, Ma X, Huang G, Ma D, Wang H. Hypomethylation and decreased expression of BRG1 in the myocardium of patients with congenital heart disease. Birth Defects Res. 2017;109:1183–1195. doi: 10.1002/bdr2.1053. [DOI] [PubMed] [Google Scholar]
- 12.Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(e47) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11(237) doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff K, Lemme M, Kahlert AK, Runde K, Audain E, Schuster D, Scheewe J, Attmann T, Pickardt T, Caliebe A, et al. DNA methylation profiling allows for characterization of atrial and ventricular cardiac tissues and hiPSC-CMs. Clin Epigenetics. 2019;11(89) doi: 10.1186/s13148-019-0679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins-Nakai R, McLaughlin P. How congenital heart disease originates in fetal life. Cardiol Clin. 2002;20:367–383. doi: 10.1016/s0733-8651(02)00008-5. v-vi. [DOI] [PubMed] [Google Scholar]
- 21.Sheng W, Qian Y, Zhang P, Wu Y, Wang H, Ma X, Chen L, Ma D, Huang G. Association of promoter methylation statuses of congenital heart defect candidate genes with Tetralogy of Fallot. J Transl Med. 2014;12(31) doi: 10.1186/1479-5876-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng W, Chen L, Wang H, Ma X, Ma D, Huang G. CpG island shore methylation of ZFPM2 is identified in tetralogy of fallot samples. Pediatr Res. 2016;80:151–158. doi: 10.1038/pr.2016.42. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Ma X, Wang H, Ma D, Huang G. Elevated methylation of the RXRA promoter region may be responsible for its downregulated expression in the myocardium of patients with TOF. Pediatr Res. 2014;75:588–594. doi: 10.1038/pr.2014.17. [DOI] [PubMed] [Google Scholar]
- 24.Grunert M, Dorn C, Cui H, Dunkel I, Schulz K, Schoenhals S, Sun W, Berger F, Chen W, Sperling SR. Comparative DNA methylation and gene expression analysis identifies novel genes for structural congenital heart diseases. Cardiovasc Res. 2016;112:464–477. doi: 10.1093/cvr/cvw195. [DOI] [PubMed] [Google Scholar]
- 25.Serra-Juhé C, Cuscó I, Homs A, Flores R, Torán N, Pérez-Jurado LA. DNA methylation abnormalities in congenital heart disease. Epigenetics. 2015;10:167–177. doi: 10.1080/15592294.2014.998536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obermann-Borst SA, Van Driel LM, Helbing WA, de Jonge R, Wildhagen MF, Steegers EA, Steegers-Theunissen RP. Congenital heart defects and biomarkers of methylation in children: A case-control study. Eur J Clin Invest. 2011;41:143–150. doi: 10.1111/j.1365-2362.2010.02388.x. [DOI] [PubMed] [Google Scholar]
- 27.Chowdhury S, Cleves MA, Macleod SL, James SJ, Zhao W, Hobbs CA. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91:69–76. doi: 10.1002/bdra.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 29.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 31.Schlesinger F, Smith AD, Gingeras TR, Hannon GJ, Hodges E. De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements. Genome Res. 2013;23:1601–1614. doi: 10.1101/gr.157271.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573:281–286. doi: 10.1038/s41586-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Bronson RT, Klaman LD, Hampton TG, Wang JF, Green PJ, Magnuson T, Douglas PS, Morgan JP, Neel BG. Mice mutant for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet. 2000;24:296–299. doi: 10.1038/73528. [DOI] [PubMed] [Google Scholar]
- 34.Taparia S, Gelineau-van Waes J, Rosenquist TH, Finnell RH. Importance of folate-homocysteine homeostasis during early embryonic development. Clin Chem Lab Med. 2007;45:1717–1727. doi: 10.1515/CCLM.2007.345. [DOI] [PubMed] [Google Scholar]
- 35.MacGrogan D, D'Amato G, Travisano S, Martinez-Poveda B, Luxán G, Del Monte-Nieto G, Papoutsi T, Sbroggio M, Bou V, Gomez-Del Arco P, et al. Sequential ligand-dependent notch signaling activation regulates valve primordium formation and morphogenesis. Circ Res. 2016;118:1480–1497. doi: 10.1161/CIRCRESAHA.115.308077. [DOI] [PubMed] [Google Scholar]
- 36.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17:2886–2893. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortensen SA, Skov LL, Kjaer-Sorensen K, Hansen AG, Hansen S, Dagnæs-Hansen F, Jensenius JC, Oxvig C, Thiel S, Degn SE. Endogenous natural complement inhibitor regulates cardiac development. J Immunol. 2017;198:3118–3126. doi: 10.4049/jimmunol.1601958. [DOI] [PubMed] [Google Scholar]
- 38.Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26:1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the figshare database (https://doi.org/10.6084/m9.figshare.14130173).