Figure 1. Cytological and transcriptomic analysis of a key miRNA/target regulon in the larval ventral nerve cord (VNC).
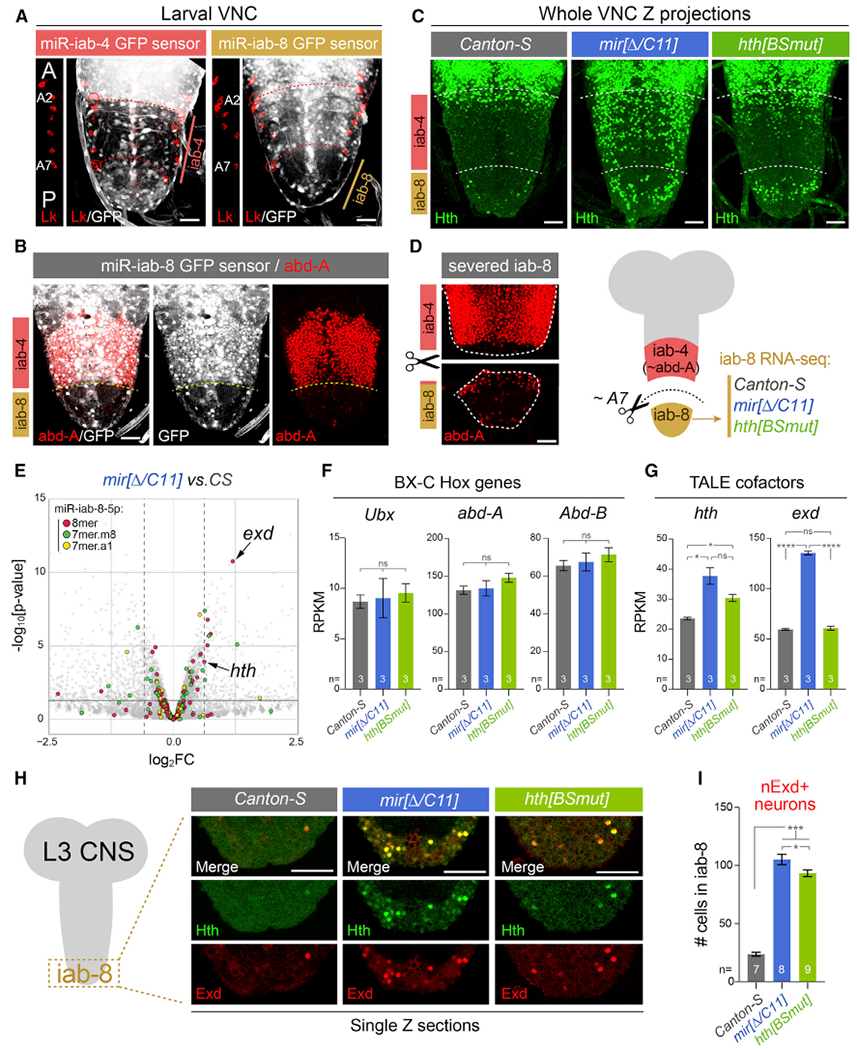
All stainings depict third instar larval VNC.
(A) The Bithorax Complex (BX-C) miRNA locus yields mir-iab-4 and mir-iab-8 from opposite strands. Their activity is associated with repression of ubiquitously transcribed tub-GFP-miRNA sensors, revealing distinct domains in abdominal segments, whose registers are marked by segmental marker Leukokinin (Lk; red).This places miR-iab-4-5p activity in segments A2–A7 and miR-iab-8-5p in A8–A9. A portion of the Lk staining is shown to the left of each image to reference the segment locations.
(B) The posterior limit of abd-A (A7a/p) corresponds to the miR-iab-4/miR-iab-8 border.
(C) Homothorax (Hth) is largely absent throughout the abdominal VNC in wild-type flies but is derepressed in mir-iab-4/8 and hth[BSmut] mutants, most overt within the iab-8 domain.
(D) Validation of dissection strategy to prospectively isolate A8–A9 VNC domain (iab-8 region) for transcriptomics. Stainings show two pieces of an individual VNC, which were imaged separately for this montage.
(E) Volcano plot of transcripts >1 reads per kilobase of transcript, per million mapped reads (RPKM) showing that TALE cofactors hth and exd are among the most significantly derepressed miR-iab-8-5p targets in the BX-C miRNA mutant VNC; most other direct targets were unaffected. Black horizontal line demarcates the 0.05 cutoff for p value; dotted vertical lines indicate the 1.5 cutoff for fold change.
(F and G) Comparison of BX-C Hox genes and TALE cofactors expression in wild-type, miRNA mutant, and hth[BSmut] iab-8 domain VNCs. exd is derepressed in the miRNA mutant, but not in hth[BSmut].
(H) Single-plane confocal images demonstrating nuclear colocalization of Hth and Exd in the iab-8 domain of mir-iab-4/8 and hth[BSmut] VNCs.
(I) Quantification of ectopic nuclear Exd cells in the iab-8 domain of VNCs of the indicated genotypes.
Statistical significance was evaluated using unpaired t test with Welch’s correction (F and G) and Mann-Whitney non-parametric test (I). Three biological replicates per genotype were used for transcriptome analysis shown in (E)–(G). *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, not significant. Error bars, SEM. Scale bars, 25 μm (A–D); 40 μm (H). A, anterior; p, posterior.
