Figure 3.
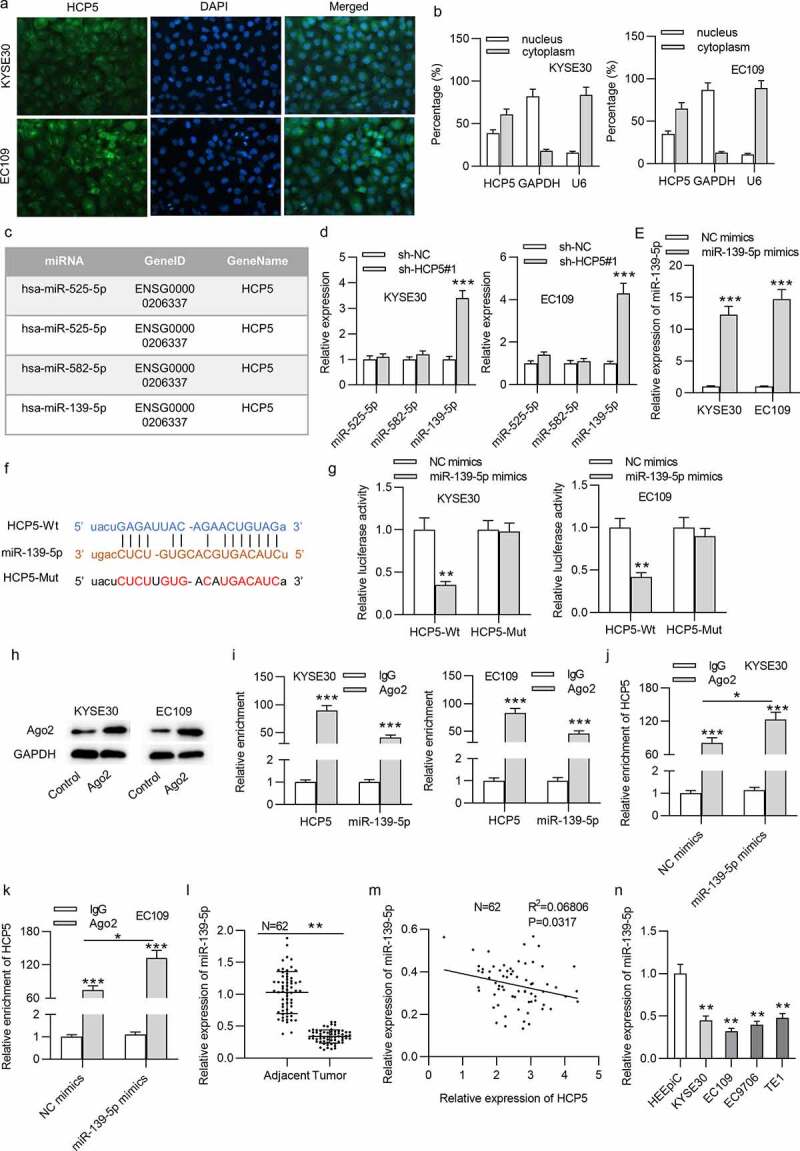
HCP5 interacts with miR-139-5p in KYSE30 and EC109 cells
(a,b) The cellular distribution of HCP5 in KYSE30 and EC109 cells was confirmed by FISH and subcellular fraction assays. (c) HCP5-binding miRNAs were predicted by starBase. (d) The expression levels of predicted miRNAs in sh-HCP5#1-treated KYSE30 and EC109 cells were detected by qRT-PCR. (e) The efficiency of miR-139-5p overexpression in KYSE30 and EC109 cells was tested with qRT-PCR. (f) The binding site of HCP5 and miR-139-5p was predicted from the starBase online database. (g) Relative luciferase activities of HCP5-Wt and HCP5-Mut plasmids in KYSE30 and EC109 were evaluated by luciferase reporter assay. (h) The effectiveness of anti-Ago2 has been verified by the western blotting. (i) RIP assay was performed to reveal the relative enrichment of miR-139-5p and HCP5 in Ago2-precipitated products in KYSE30 and EC109 cells. (j,k) RIP assay was performed to reveal the relative enrichment of HCP5 in Ago2-precipitated products in KYSE30 and EC109 cells transfected with NC mimics and miR-139-5p mimics. (l) MiR-139-5p expression in 62 paired ESCC tissues and corresponding nontumor tissues was detected by qRT-PCR. (m) Expression correlation of HCP5 and miR-139-5p in 62 ESCC tissues was analyzed by Pearson’s correlation analysis. (n) MiR-139-5p expression in ESCC cells (KYSE30, EC9706, EC109, TE1) and normal esophageal epithelial cell line HEEpiC was detected by qRT-PCR. *p < 0.05, **p < 0.01, ***p < 0.001.
