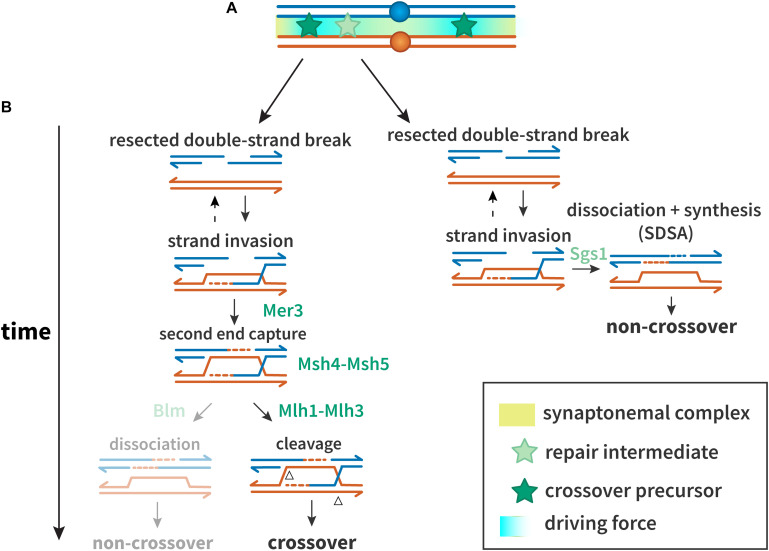
FIGURE 1.
Proteins and mechanisms of crossover interference. (A) A driving force (teal), potentially protein aggregation or mechanical stress, results in designation of crossover precursors (stars, dark green) at spaced intervals. Repair intermediates that do not experience sufficient driving force (stars, light green) are not so designated, and become non-crossovers. (B) Interference at the DNA level. Crossover precursors are processed through the class I crossover pathway, involving the ZMM proteins. Mer3 promotes second end capture, and Msh4–Msh5 clamp to and stabilize branched molecules to prevent their dissociation and to recruit Msh1–3. Intermediates are resolved by Mlh1–Mlh3 to generate crossovers. Non-crossovers in yeast are thought to be exclusively processed through the synthesis-dependent strand annealing (SDSA) pathway, promoted by Sgs1. In other organisms, the Sgs1 ortholog Blm may also promote formation of non-crossovers after second end capture.