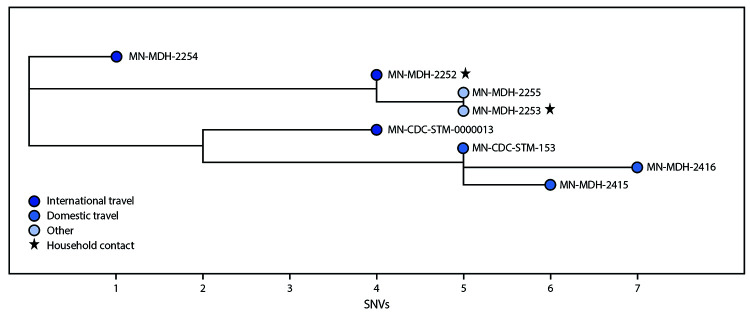
FIGURE.
Phylogenetic tree* showing genetic distance† between SARS-CoV-2–positive specimens with the B.1.1.7 variant (n = 8) and exposure histories related to travel,§ household contacts, and others in the community
Abbreviation: SNV = single nucleotide variant.
* Phylogenetic tree created using Interactive Tree of Life (version 5.7; European Molecular Biology Laboratory). https://itol.embl.de/
† MN-MDH-2253 and MN-MDH-2255 were identical, and MN-MDH-2252 was within one SNV. MN-MDH-2252 and MN-MDH-2253 were collected from persons who were household contacts. Two specimens from international travelers, MN-MDH-2254 and MN-CDC-STM-0000013, did not have sequences similar to those identified in Minnesota.
§ International and domestic travel occurred during the 14 days before illness onset or specimen collection, including onset of illness in persons who received positive SARS-CoV-2 test results while away from Minnesota. MN-CDC-STM-153, MN-MDH-2416, and MN-MDH-2415 were clustered together within one to three SNVs. These three specimens were from persons who reported travel to California in the 14 days before illness onset or specimen collection, including one who received a positive test result while in California and isolated there before returning to Minnesota.