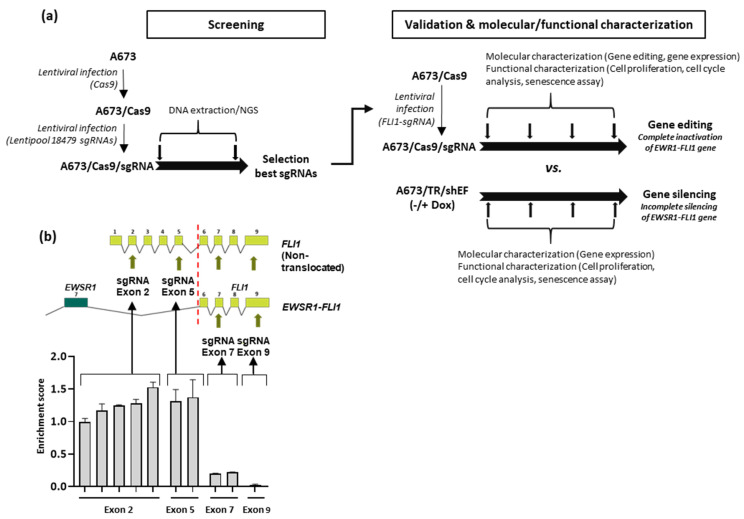
Figure 1.
Selection of gRNAs targeting EWSR1–FLI1 and analysis of gene editing evolution. (a) Experimental design: Ewing sarcoma cell line A673 expressing Cas9 protein (A673/Cas9) were generated by lentiviral infection. After clonal selection, A673/Cas9 cells were infected with a multiplex lentiviral CRISPR library of 18,479 different sgRNAs targeting 1983 transcription factors including ten gRNAs targeting FLI1. After this screening phase, two gRNAs were selected for functional and molecular characterization. A673/Cas9 were infected with lentiviral sgRNAs to generate A673/Cas9/sgRNA cells and then maintained in continuous growth to assess gene editing, cell proliferation, senescence, and studies of mRNA and protein expression at different time points. A673/TR/shEF, which expresses a specific EWSR1–FLI1 shRNA upon doxycycline stimulation, were cultured and analyzed in a similar way. The results obtained upon gene editing and gene silencing were then compared. (b) Schematic representation of native FLI1 and EWSR1–FLI1 fusion genes, location of FLI1 gRNAs, and enrichment scores obtained for each gRNA in the CRISPR screening assay (mean ± SD of two independent experiments).