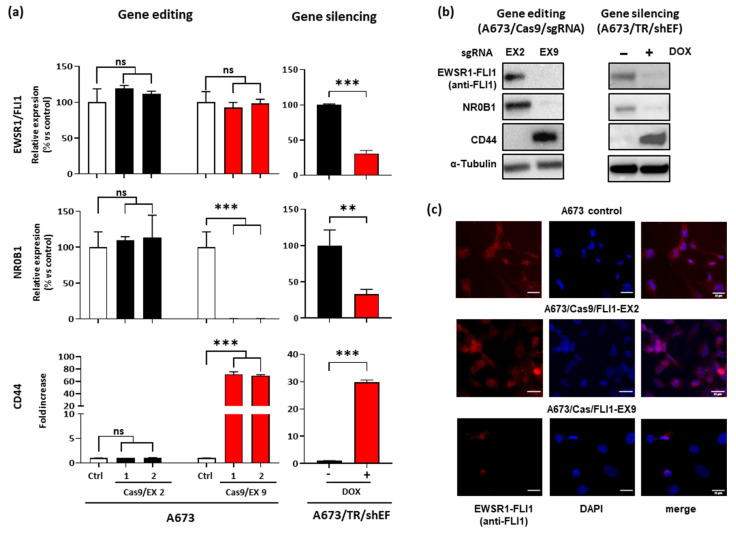
Figure 2.
Effect of EWSR1–FLI1 gene editing on EWSR1–FLI1 expression and EWSR1–FLI1 target genes. (a) EWSR1–FLI1, NR0B1, and CD44 mRNA levels were quantified by RT-qPCR in A673/Cas9 cell lines infected with sgRNA vectors targeting exon 2 and exon 9. The graphs represent the results of two independent experiments performed with MOI = 1 (1) and MOI = 5 (2). A673/sgRNA was used as the control (Ctrl). A673/TR/shEF cells were stimulated with doxycycline (1 µg/mL) for 72 h to induce the expression of the EWSR1–FLI1-specific shRNA. Data shown are the mean ± SD of experiments conducted in triplicate (ns, not significant; ** p < 0.01; *** p < 0.001; Student’s t-test). (b) EWSR1–FLI1, NR0B1, and CD44 protein levels were detected by Western blot. β-Tubulin was used as a control for loading and transferring. (c) Immunostaining of cells with anti-FLI1 antibody. Intense nuclear staining for EWSR1–FLI1 (red fluorescence colocalized with DAPI staining) was observed in both the A673 control cell line and A673/Cas9/FLI1-EX2, while it was undetectable in the majority of the A673/Cas9/FLI1-EX9 cells.