Fig. 5. PINK1-dependent formation of LC3-positive autophagosomes precedes rapid lysosome fusion.
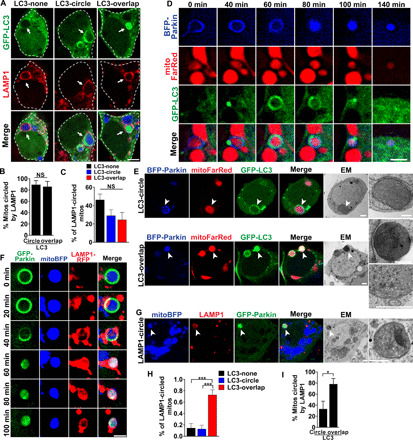
(A) Drp1KO neurons expressing GFP-LC3 (green) and mitoBFP (blue) treated with 100 nM bafilomycin for 4 hours and stained for LAMP1 (lysosomes, red). Scale bar, 4 μm. (B) Most mitochondria with circling or overlapping patterns of GFP-LC3 were circled by LAMP1. n = 8 coverslips per group, 4 to 7 neurons per coverslip, three experiments. (C) Among LAMP1-circled mitochondria, ≈50% did not have LC3 accumulation (diffuse LC3); others were circled by GFP-LC3 (LC3-circle) or had GFP-LC3 overlapping the mitochondrial matrix (LC3-overlap). n = 8 coverslips per group, n = 6 to 11 cells per coverslip, three independent experiments. (D) Drp1KO neurons coexpressing GFP-LC3, mitoFarRed, and BFP-Parkin. GFP-LC3 accumulates in a distinct punctum on the mitochondrion with OM-Parkin, before dispersing to encircle the mitochondrion. BFP-Parkin then transitions to the overlapping pattern, and LC3 reaccumulates into a punctum. Scale bar, 2 μm. (E) CLEM shows ultrastructure of Parkin-positive mitochondria with distinct patterns of GFP-LC3. Mitochondria with LC3-circle are surrounded by an isolation membrane (top). LC3-overlap mitochondrion (bottom) is electron-dense but intact, with packed cristae (see insets). Bottom inset is the same structure from a different section. Scale bars, 1 μm. (F) Drp1KO neurons coexpressing GFP-Parkin, LAMP1-RFP, and mitoBFP. Images show formation of a LAMP1 ring, indicating lysosomal fusion, concurrent with Parkin shifting from OM to overlapping. Scale bar, 2 μm. (G) Overlapping-Parkin mitochondria circled by LAMP1 are mitolysosomes by ultrastructure. Scale bars, 1 μm. (H) Drp1KO;PINK1KO neurons expressing GFP-LC3 and mitoBFP treated with bafilomycin for 4 hours and stained for LAMP1. Among LAMP1-circled mitochondria, ≈80% had LC3-overlap, and a few were either circled or lacked GFP-LC3 accumulation. n = 9 coverslips per group, 5 to 12 neurons per coverslip, two experiments. (I) Most mitochondria with LC3-overlap were circled by LAMP1. n = 6 coverslips per group, 5 to 12 neurons per coverslip, three experiments. *P < 0.05 and ***P < 0.001 by t test (B and I) or one-way ANOVA with Tukey’s multiple comparisons test (C and H).
