Figure 6. Mimicking HNRNPM-dependent linear-splicing events in cells inhibits exon inclusion and cell growth.
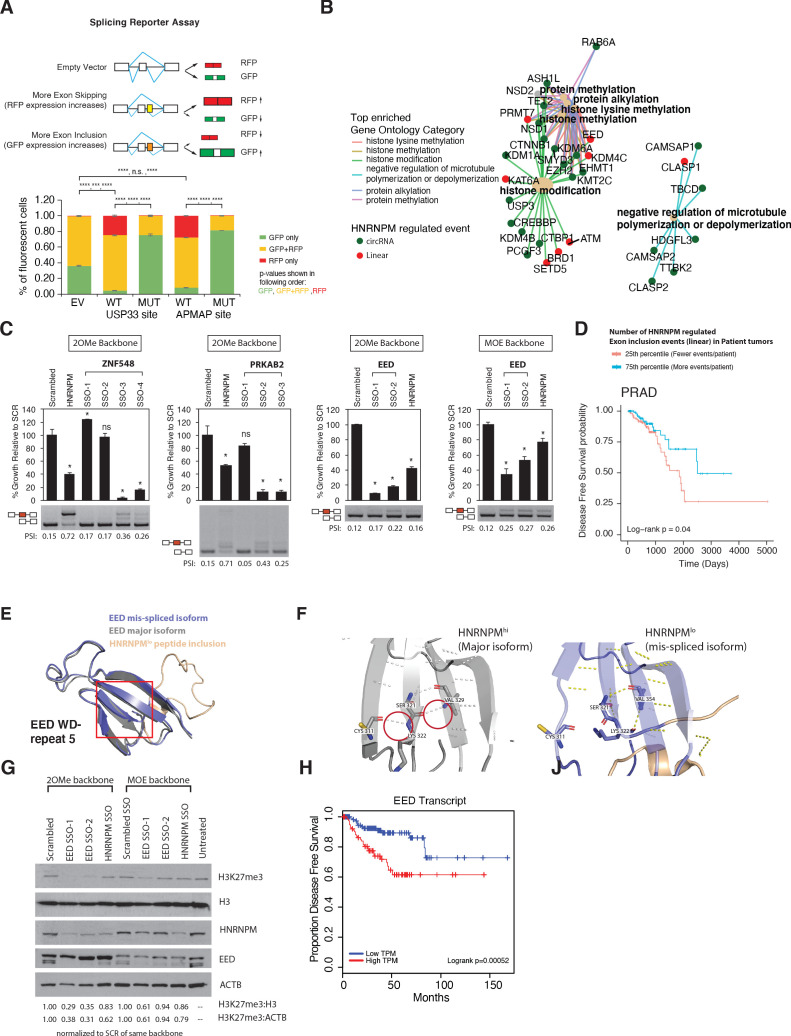
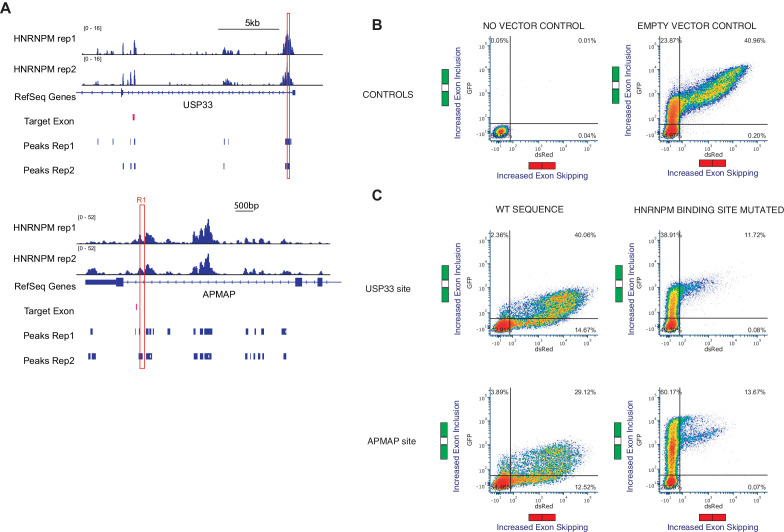
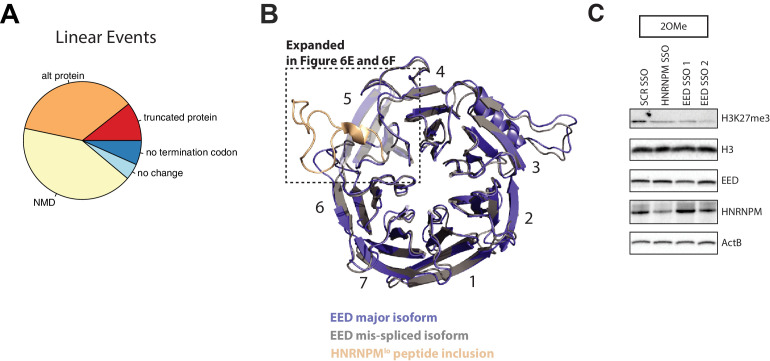
(A, top panel) Schematic showing outcomes of splicing reporter assay. Wildtype (WT) or mutant (MUT) HNRNPM-binding sites at the USP33 or APMAP genes and identified by eCLIP were cloned into a splicing reporter. The percentages of green fluorescent protein (GFP) (indicating exon inclusion) and dsRed fluorescent proteins (RFP) (indicting exon skipping) single- (green and red bars) and double-positive cells (yellow bars) out of all fluorescent cells are shown in a barplot in the lower panel. Shown are the mean and standard deviation of three experiments. Student’s t-test was used to calculate p-values between the indicated groups. Ns: not significant; ** p<0.01; **** p<0.0001. (B) Gene ontology analysis of transcripts that are either changed in expression or mis-spliced upon HNRNPM knockdown. Both mis-spliced circular (green dots) and linear (red dots) RNAs are shown. Top enriched gene ontology categories (yellow dots) and the genes present in each category are indicated with colored lines. (C) Cell proliferation and exon inclusion outcomes in splice-switching-antisense-oligonucleotides (SSO)-treated cells as compared to scrambled SSO-treated cells. For proliferation assays, growth is shown relative to cells treated with the scrambled non-targeting SSO. The mean and standard deviation of three biological replicates are shown. p-values for growth were determined using Student’s t-test. *p<0.05. A representative splicing gel together with the PSI of the HNRNPM-regulated isoforms are shown below each barplot. As controls, a HNRNPM targeting SSO is also included (lane 2). For each gene (ZNF548, PRKAB2, and EED), multiple SSOs targeting different sequences were used. ZNF548 SSO-1, ZNF548 SSO-2, and PRKAB SSO-1 are control SSOs that do not induce inclusion events. For the EED event, both 2-O-methyl (2OMe) and 2-methoxy-ethyl (MOE)-based SSO backbone chemistries were used to control for potential toxicities due to SSO transfection. SSO backbone chemistries used are indicated at the top of each panel. (D) Disease-free survival plot of The Cancer Genome Atlas (TCGA) prostate adenocarcinoma (PRAD) patients, stratified by the total number of events per patient where the PSI of a given HNRNPM-regulated exon exceeds that of the median PSI within the patient population. Patients with more exon inclusion events (top 75th percentile) are shown in blue, whereas patients with less exon inclusion events (lower 25th percentile) are shown in red. (E) WD40 domain 5 of EED when the HNRNPM-regulated exon 10 is included. Structure of the HNRNPM-regulated WD40 domain in WT EED (gray), superimposed on the predicted structure of the HNRNPM-dependent EED isoform (blue). The new peptide generated by the splicing event is depicted in orange. (F) Closeup view of hydrogen bonding interactions that stabilize the WD40 beta sheet in either WT or the new EED isoform. (G) Western blot of H3K27me3 (EED target), HNRNPM, and EED upon treatment with the scrambled, HNRNPM-targeting SSO (HNRNPM) or SSOs that promote the inclusion of EED exon 10 (EED SSO-1 and SSO-2). Backbone chemistry of the SSOs used is indicated. Untreated cells are included as controls. HNRNPM. Shown also are histone 3 and actin B loading controls. (H) Disease-free survival curves of prostate cancer patients when expression in transcripts per million (TPM) of EED is low (blue) or high (red).