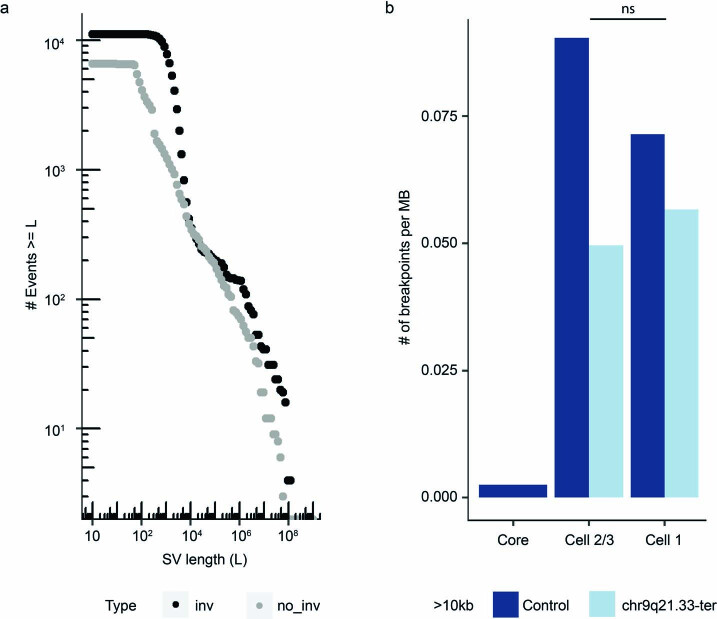
Extended Data Fig. 5. Replication and collective missegregation of Chr.9q21.33-ter did not result in chromothripsis.
a, Length distribution of inverted and non-inverted structural variants (SVs) in four selected single cells of the dataset presented in Fig. 3 (cell 1 (5 copies of a chr9.q fragment)), cell 2 and 3 (1 copy) and a cell with the core karyotype (diploid)). The enrichment of short-range inverted SVs can be explained by the WGA reaction. A similar count in inverted and non-inverted SVs was observed for SVs with a length ≥10 kb. b, The frequency of SVs (length ≥ 10 kb) on all control chromosomes and on the missegregated chr.9q21.33-ter fragment. No enrichment of SVs was observed in the chr.9q21.33-ter region for Cell No. 1 vs. Cell No. 2/3 which suggests that chromothripsis did not occur (p = 1, two-sided Fisher’s exact test).