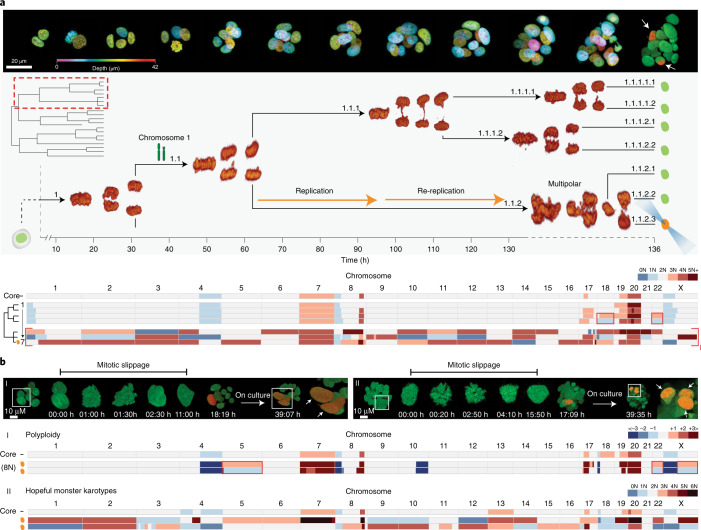
Fig. 4. Genome duplication events are substrates for hopeful monster karyotypes.
a, 3D Live-Seq dataset of a PDTO-9 organoid consisting of 19 cells (95% recovery). Top: representative stills of the growing PDTO-9 structure with nuclei coded in a false color by depth. Photoconverted cells are indicated in the final still (white arrows). One photoconverted cell was mapped to branch 1.1 of the mitotic tree, as highlighted (dashed red box) in the full tree structure at the top left of the middle panel and enlarged in the main image of the middle panel, with 3D-rendered stills of each anaphase. The onset of anaphase is indicated by arrowheads in relation to the time axis. Bottom: karyotype heatmap of all cells mapped to branch 1.1 (all cells share a loss of chromosome 1p; chromosome cartoon) and a reference core karyotype of the imaged PDTO. Three cells, including the photoconverted cell, showed reciprocal hopeful monster karyotypes (I) and were mapped to tripolar division 1.1.2 using the photoconversion reference landmark. Cell 1.1.2 underwent re-replication before anaphase, as indicated by the difference in ploidy between branches 1.1.1 and 1.1.2. b, Daughter cells post-mitotic slippage maintain polyploidy of the genome-duplicated ancestor. Lineage I: representative imaging stills (top) of a binucleated cell undergoing mitotic slippage. A PDTO with a photoconverted slippage cell was cultured overnight. The final still shows two photoconverted daughter cells (white arrows) with a similar chromatin mass. Bottom: karyotype heatmap of both daughter cells and the remaining PDTO cells (core). Daughter cells showed polyploidy (8 N) but roughly maintained the core karyotype. Heatmap colour coding indicates deviations with respect to the ploidy of the core karyotype (2N) or daughter cells (8N). The red boxes indicate reciprocal gains and losses. Lineage II: as described for lineage I, but showing a multipolar spindle defect after mitotic slippage resulting in hopeful monster karyotypes among three daughter cells (white arrows). Sequencing data were obtained from two out of three daughter cells and display gross genome-wide karyotype alterations relative to the core karyotype.