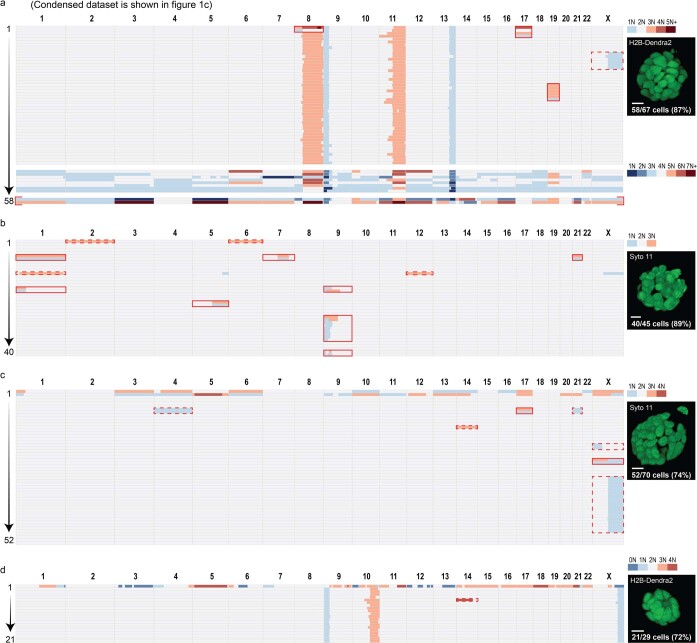
Extended Data Fig. 2. Full and additional whole population PDTO sequencing datasets of PDTO-19b.
a, Full PDTO-19b dataset of experiment shown in Fig. 1c. Karyotype heatmap showing 58 cells derived from a clonal PDTO-19b structure expressing transgenic H2B-Dendra2 and consisting of 67 cells (87% recovery). Reciprocal gains and losses are indicated with red boxes. Dashed boxes indicate CNA events where a reciprocal loss or gain is missing. Sub-chromosomal CNAs were counted as events when represented in more than one cell. The bottom panel shows a population of polyploid cells with large deviations from the core karyotype (hopeful monsters). Two cells show reciprocal gains and losses across their genome (red brackets). Scalebar is 10μm. b, As in a. Genetically unmodified PDTO-19b organoid. Cells were stained with Syto 11 to allow single cell picking. 40 out of 45 cells provided quality sequence (89% recovery). c, As in a. Genetically unmodified PDTO-19b organoid. Cells were stained with Syto 11 to allow single cell picking. 52 out of 70 cells provided quality sequence (74% recovery). d, As in a. 21 out of 29 cells were sequenced (72% recovery). The previously reported TP53 mutation (chr 17: 7577121 G > A) for PDTO-19b [van de Wetering, 2016] was confirmed in all sequenced clonal organoids.