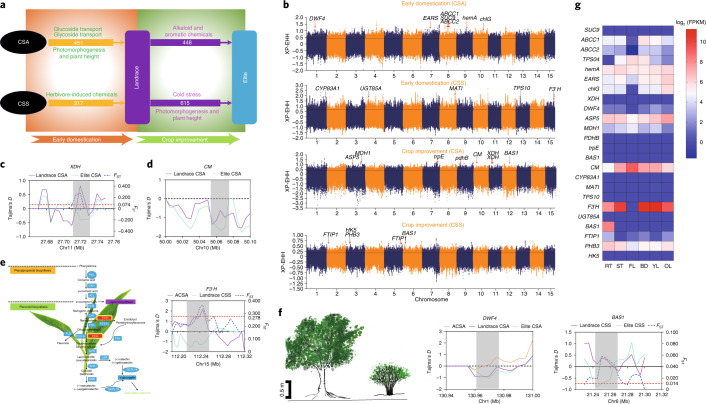
Fig. 5. Signatures of artificial selection and evidence of parallel domestication in CSA and CSS.
a, A proposed road map of parallel domestication in CSA and CSS. Early domestication involved genes related to glucoside and glycoside transport and photomorphogenesis and plant height in CSA and herbivore-induced chemicals in CSS. The improvement mainly focused on genes related to alkaloid and aromatic chemicals in CSA and genes related to cold stress and photomorphogenesis and plant height in CSS. The number of artificially selected genes are labeled in each domestication process. b, Genome-wide distribution of selective-sweep signals identified based on cross-population extended haplotype homozygosity (XP-EHH). Genes with important functions that are linked to sweeps are highlighted in Manhattan plots. Genes that were involved in early domestication were identified based on a comparison between CSA and CSS landraces, and ACSA, while genes under improvement were selected based on a comparison between CSA and CSS elite, and CSA and CSS landraces. c, Signals of artificial selection in the XDH gene. Purple and green solid lines indicate Tajima’s D statistics in CSA landraces and CSA elite cultivars, respectively. The blue dashed line indicates the fixation index (FST) between CSA landrace and elite populations, while the red dashed line is the threshold of the top 5% FST. d, Signals of artificial selection in the CM (chorismate mutase) gene. Purple and green solid lines indicate Tajima’s D statistics in CSA landraces and CSA elite cultivars, respectively. e, Signals of artificial selection in the F3′H gene. Left, F3′H is one of the key genes in catechin biosynthetic pathways. Right, signals of artificial selection in this gene. f, Signals of artificial selection in BAS1 and DWF4 genes, related to plant height. The leftmost panel shows different morphological features in tea accessions. In contrast to ACSA, CCSA and CCSS are featured with decreased plant height, with most CCSA being small trees or semi-shrubs and CCSS being shrubs. Scale bar, 0.5 m. Middle and rightmost panels show signals of artificial selection in BAS1 and DWF4. g, Expression of artificially selected genes in the six tissues examined, including root (RT), stem (ST), flower (FL), bud (BD), young leaf (YL) and old leaf (OL).