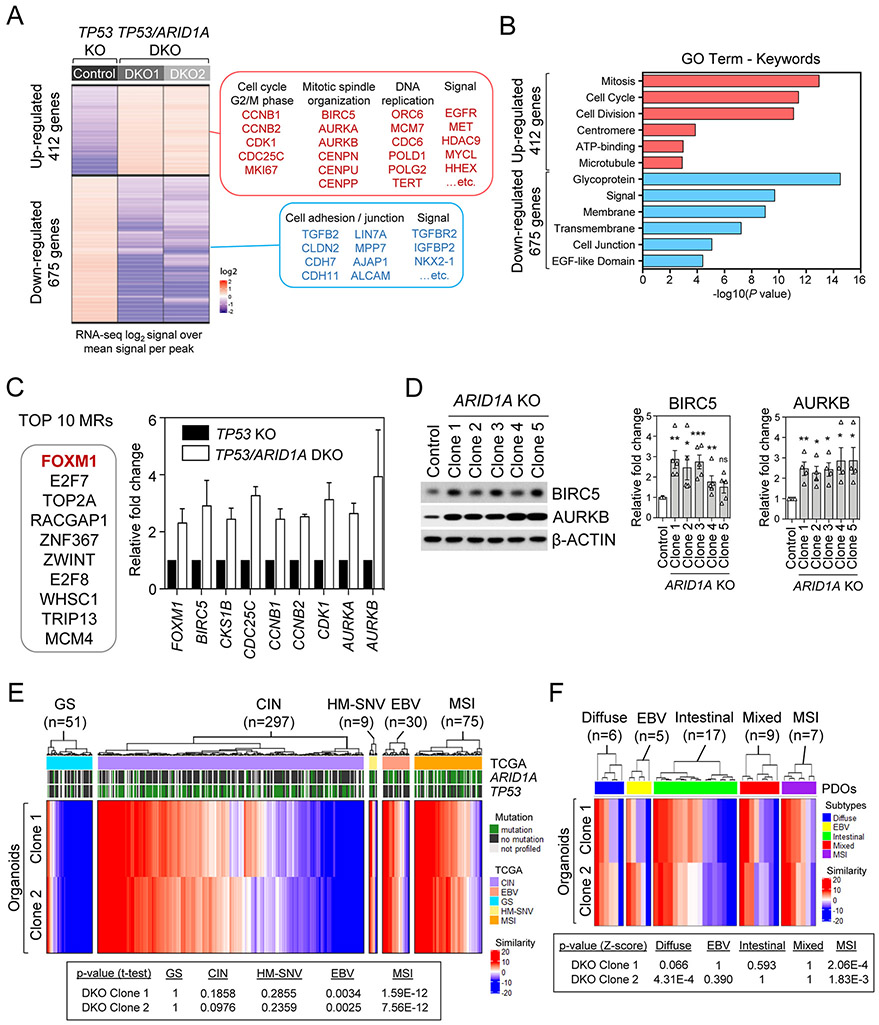
Figure 5. ARID1A loss-associated gene master regulatory modules identify a FOXM1/BIRC5 node and recapitulate TCGA MSI and EBV human gastric cancers.
A, Heatmap of significant differentially expressed genes with at least 2-fold change in each TP53/ARID1A DKO lines, compared with TP53 KO control line. A total of 412 up-regulated genes and 675 down-regulated genes were identified. Selected genes and signaling pathways are listed. B, Gene ontology analysis identified top key terms significantly associated with transcriptional profiles in TP53/ARID1A DKO organoids. C, Top 10 master regulators from ARACNe and VIPER prediction that were activated in TP53/ARID1A DKO organoids versus control TP53 KO are reported. Several FOXM1 targets, including BIRC5, CKS1B, CDC25C, CCNB1, CCNB2, CDK1, AURKA and AURKB were significantly upregulated in ARID1A-deficient cells. D, Western immunoblotting analysis demonstrated that FOXM1 targets, BIRC5 and AURKB, were upregulated in TP53/ARID1A DKO organoids. Quantification of BIRC5 and AURKB expression from independent experiments (N>3) was shown. Dots indicate independent experiments. The horizontal bar indicates mean. The error bar represents SEM. E, Comparison of master transcriptional regulators in ARID1A KO organoids to TCGA STAD gastric cancer patient cases indicated significant similarities between organoids and TCGA MSI and EBV subtypes. The p-value computed by t-test (one sample) with the alternative hypothesis of true mean of the similarity score is greater than zero. Red and blue colors indicate high and low similarity concurrence, respectively. F, Comparison of master transcriptional regulators in ARID1A-deficient organoids to gastric cancer patient-derived organoids (PDOs) indicated significant similarities between engineered TP53/ARID1A DKO organoids and MSI subtype PDOs.