Figure 5.
Effect of paternal Dex injection on embryonic offspring long RNA transcriptome
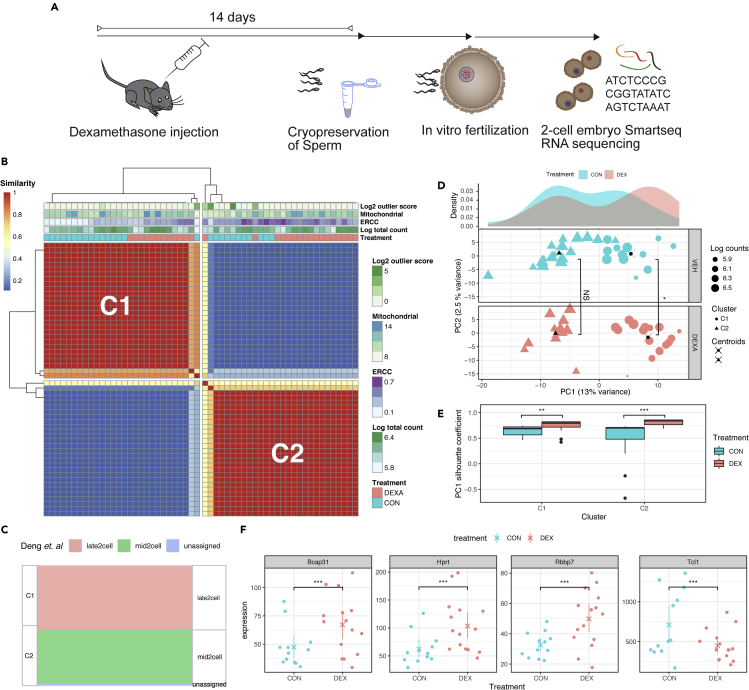
(A) Experimental design depicting timeline between injection, sperm harvest, in vitro fertilization, and Smartseq2 sequencing at 2-cell stage.
(B) Consensus matrix representing the similarity between cells as reported by SC3. Similarity 0 indicates that a given pair of embryos was never assigned to the same cluster, whereas similarity 1 means that a pair of embryos was always assigned to the same cluster.
(C) Sankey diagram showing projection of the obtained clusters (C1 and C2) into clusters reported by Deng et al. for single cells obtained from 2-cell embryos.
(D) Principal component analysis of two-cell embryos gene expression. The top panel indicates the density of 2-cell embryos along PC1 grouped by condition; control (red) and Dex treatment (blue). The two bottom panels show the distribution of 2-cell embryos across PC1 and PC2 for control (red) and treated (blue) groups. The cluster membership of each embryo is denoted by the point shapes (C1 cycles; C2 triangles), and the centroids of each cluster are indicated with a black symbol overlaid with an x. Wilcox tests were performed to assess differences on PC1 values of C1 and C2 clusters between the treated and control groups. NS denotes nonsignificant change for C2 cluster, while ∗ indicates a significant difference for C1 cluster (p value< 0.05).
(E) Silhouette coefficient comparison between treatment and control, statistical significance was assessed with Wilcox test (∗∗ p value < 0.01; ∗∗∗ p value < 0.005)
(F) Selection of differentially expressed genes as determined by Monocle within C1 corresponding to late 2-cell embryo stage (∗∗∗ adjusted p value < 0.005). Error bars display 95% nonparametric confidence intervals.