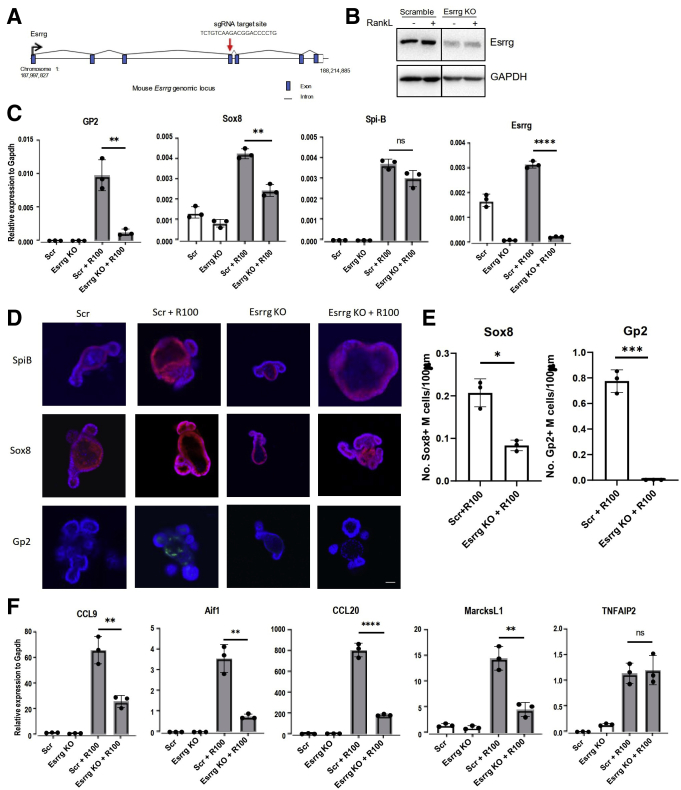
Figure 5.
Abolition of Esrrg impairs Sox8 activation and the functional maturation of M cells. (A) Schematic representation of Esrrg KO design by CRISPR-Cas9 genome editing in mouse intestinal organoids. Exon, intron, and genomic position are indicated. (B) Esrrg protein expression in Esrrg KO and Scrambled cells generated by lentiCRISPR v2 genome editing in C57BL/6JRj intestinal organoids. Organoid lysates were analyzed by Western blot. (C) qPCR analysis of M-cell–associated genes expressed in Scrambled and Esrrg KO stimulated by RankL for 4 days. (D) Immunostaining images for Spi-B (red), Sox8 (red), and GP2 (green) in Scrambled organoids with and without 100 ng RankL for 4 days and Esrrg KO with and without 100 ng RankL. Scale bars: 100 μm. (E) The number of GP2+ M cells and Sox8+ M cells per length of epithelium of organoids was compared between Scr+R100– and Esrrg KO+R100–treated organoids (n = 3). Images are representative of 3 independent experiments. (F) qPCR analysis of early markers of M-cell–associated genes expressed in vitro in the presence of RankL for 4 days. Values in all are presented as the means ± SD. Unpaired 2-tailed Student t test, N = 3 independent experiments. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ∗∗∗∗P < .0005. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; sgRNA, single guide RNA.