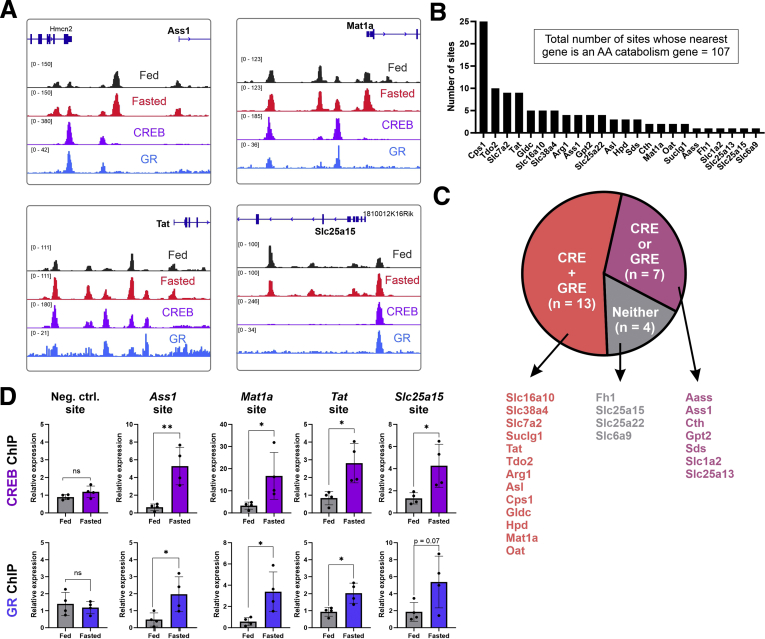
Figure 2.
CREB and GR bind fasting-induced enhancers adjacent to fasting-induced AA catabolism genes. Loci of 4 fasting-induced genes are presented. Black and red tracks represent normalized DNase accessibility of hepatic chromatin in the fed vs 24-hour fasted states, respectively. Violet and blue tracks represent normalized ChIP-seq of CREB and GR, respectively, after treatment of glucagon and corticosterone in primary mouse hepatocytes. Increased chromatin accessibility as well as evident CREB and GR binding at enhancers is observed adjacent to fasting-induced AA catabolism genes (A). Number of fasting-induced enhancers adjacent to each AA catabolism gene is depicted (7 genes with no fasting-induced enhancers are not shown) (B). Genes with adjacent fasting-induced enhancers were divided into 3 groups: genes whose proximal enhancer contain both a CRE and a GRE (n = 13), either a CRE or a GRE (n = 7), or neither (n = 4) (C). Chromatin immunoprecipitation of CREB and GR was performed on mice fasted for 24 hours and on ad libitum fed mice. CREB and GR showed increased binding at sites near fasting-induced, synergistic AA catabolism genes. Graphs represent data collected from 4 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05 (D).