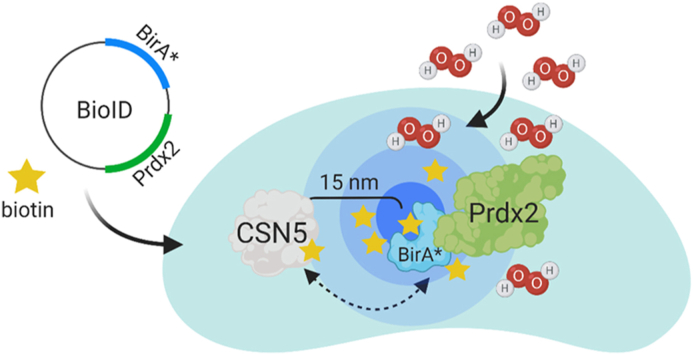
Abstract
Hydrogen peroxide (H2O2) acts as a signalling molecule by oxidising cysteine thiols in proteins. Recent evidence has established a role for cytosolic peroxiredoxins in transmitting H2O2-based oxidation to a multitude of target proteins. Moreover, it is becoming clear that peroxiredoxins fulfil their function in organised microdomains, where not all interactors are covalently bound. However, most studies aimed at identifying peroxiredoxin interactors were based on methods that only detect covalently linked partners. Here, we explore the applicability of two thiol-disulphide independent in-cell trapping methodological approaches in combination with mass spectrometry for the identification of interaction partners of peroxiredoxin 2 (Prdx2). The first is biotin-dependent proximity-labelling (BioID) with a biotin ligase A (BirA*)-fused Prdx2, which has never been applied on redox-active proteins. The second is crosslinker co-immunoprecipitation with an N-terminally His-tagged Prdx2. During the initial characterisation of the tagged Prdx2 constructs, we found that the His-tag, but not BirA*, compromises the peroxidase and signalling activities of Prdx2. Further, the Prdx2 interactors identified with each approach showed little overlap. We therefore concluded that BioID is a more reliable method than crosslinker co-immunoprecipitation. After a stringent mass spec data filtering, BioID identified 13 interactors under elevated H2O2 conditions, including subunit five of the COP9 signalosome complex (CSN5). The Prdx2:CSN5 interaction was further confirmed in a proximity ligation assay. Taken together, our results demonstrate that BioID can be used as a method for the identification of interactors of Prdxs, and that caution should be exercised when interpreting protein-protein interaction results using tagged Prdxs.
Keywords: Peroxiredoxin, Redox signalling, BioID, Proximity labelling, Protein-protein interactions, Prdx2
Graphical abstract
Highlights
-
•
BioID is a thiol-disulphide independent method that can be used to trap Prdx2 interactors.
-
•
Prdx2 interacts with subunit five of the COP9 signalosome complex (CSN5).
-
•
N-terminally His-tagged Prdx2 displays compromised peroxidase and signalling activity.
-
•
The tag attached to Prdx2 influences its interactome.
1. Introduction
H2O2 is a key redox signalling molecule that reversibly oxidises redox sensitive cysteines in proteins [1], resulting in an altered protein structure [[2], [3], [4]], function [3,5,6], or cellular location [7,8]. However, given the low reactivity of most thiols with H2O2 [9] and the presence of highly abundant and extremely efficient H2O2 scavenging peroxiredoxins (Prdx) [10,11], it is surprising to observe that H2O2 can oxidise the cysteine thiols of its target proteins. The most likely explanation is that it is the Prdxs that transfer oxidative equivalents from H2O2 to redox-sensitive proteins via a redox relay mechanism [12,13]. According to this mechanism, first the peroxidatic cysteine (CysP) of Prdx reacts with H2O2, forming a sulphenic acid (CysP-OH). Then, the thiol of the partner protein reacts with CysP-OH, followed by thiol-disulphide exchange with a vicinal thiol in the partner, if available. In a different scenario, the Prdx CysP-OH forms a disulphide bond with the resolving Cys (CysR) and the thiol of the partner protein then reacts with the disulphide within Prdx using a typical thiol-disulphide exchange mechanism.
In the cytosol of mammalian cells, the global existence of Prdx1-and Prdx2-mediated redox relays has been demonstrated [14], and especially the interaction between Prdx2 and the transcription factor STAT3 has been studied in detail in terms of its redox relay [15]. For a long time, the question of how Prdxs and their target proteins recognise each other and position themselves for the transfer of oxidative equivalents remained unclear. Recently, the membrane chaperone annexin A2 (AnxA2) was found to template the Prdx2:STAT3 interaction for a productive redox relay [16].
Most cytosolic Prdx interactors, including STAT3 and ASK1, have been found using methods such as conventional affinity pull-down methods or immunoprecipitation (IP), ‘diagonal' non-reducing vs reducing gel electrophoresis, or kinetic trapping with thioredoxin (Trx) mutants [14,15,17,18]. These methods have several limitations. Diagonal gels only identify interactors which form mixed disulphide bonds with peroxiredoxins. While this might be the prevailing mechanism by which peroxiredoxins transfer oxidative equivalents to target proteins, a two-step redox relay mediated by glutaredoxins cannot be excluded. Indeed, the sulphenic acid formed on the peroxidatic cysteine of Prdx2 has been demonstrated to form mixed disulphides with glutathione with subsequent reduction by glutaredoxins. In turn, glutaredoxins are known to be able to oxidise proteins [[19], [20], [21]]. Thioredoxins could also oxidise proteins by a similar two-step redox relay mechanism [22], though it has been demonstrated that, at least in the specific context of cytosolic 2-Cys peroxiredoxins (Prdx1 and Prdx2) in HEK293T cells, this is not the case [14]. Methods based on Trx-trapping mutants only detect Prdx interactors forming disulphide bonds that are reduced by thioredoxin, but not other oxidative modifications, such as S-glutathionylation. Finally, non-covalent interactors are also excluded from hits generated this way. In light of the discovery that peroxiredoxins form redox relays as part of organised membrane-associated complexes [16], having a method that allows the capture of thiol-independent interactors is becoming especially important.
Another source of potential caveats from these methods arises from the fact that the trapping of interaction partners occurs post cell lysis. Even though most studies take caution to prevent this by using thiol-blocking reagents, e.g., iodoacetamide (IAM), methyl methanethiosulphonate (MMTS), N-ethylmaleimide (NEM) [23], or antioxidant enzymes like catalase [24], the probability of generating false positive disulphide bonds due to oxidation during cell lysis still remains [25], with reports of MMTS actually promoting intra- and intermolecular disulphide bond formation [26]. Moreover, common immunoprecipitation protocols include the use of harsh buffer solution conditions (i.e. containing high salt and detergent concentrations) during washing steps, which renders it difficult to maintain intact protein complexes. Hence, complex components including potential scaffolding proteins that form non-covalent interactions could be easily disrupted, and therefore lost.
To overcome these limitations, a thiol-disulphide independent method that labels interacting proteins in cells pre-lysis is needed. Such a method can simply be a modified immunoprecipitation protocol that includes the crosslinking of interacting proteins with a chemical crosslinker added to the cells before lysis. This approach has been successfully implemented in numerous interactor studies, and chemical crosslinkers with different properties are available, such as DSSO [27], DSP [28], DSBU [27], and EDC [29].
The proximity-dependent labelling method BioID also fulfils these criteria and can therefore serve as an additional approach to investigate peroxiredoxin interactomes. BioID, as a protein-protein interaction (PPI) identification method, utilises the mutant of the promiscuous biotin ligase (BirA*) from E. coli fused to the protein of interest (the “bait”) [30]. Upon expression in cells, the fusion protein constantly activates free biotin in the presence of ATP, which then biotinylates the proteins in its vicinity (estimated radius ~10–15 nm [31]). These biotinylated proteins can be considered as interaction partners of the bait protein and include weak and transient interactors. Another advantage of this technique lies in its dependency on the biotin-avidin interaction. As this is the interaction with the highest affinity known in nature (KD = 10−14 M [32]), it can withstand stringent washing conditions necessary to dissociate unlabeled proteins during the immunoprecipitation and mass spec sample preparation better and hence yield more specific results than other methods. What makes this method especially suitable for redox biology is that, unlike some other alternative proximity-labelling techniques based on engineered ascorbate peroxidase tagging [33], it does not require the addition of external H2O2 to induce biotinylation and can therefore be used to identify Prdx interactors specifically occurring under elevated H2O2 conditions that, due to the nature of BioID, would still be independent of the redox state of thiols. BioID has been successfully implemented in numerous studies to identify novel PPIs, such as peptidyl-prolyl cis-trans isomerase FKBP4, a novel PI3K-Akt-mTOR interactor [34]; several prohibitin-2 associated mitochondrial intermembrane space proteins [35], and multiple human centriolar satellite interactors [36], yet never in the field of redox biology.
In this study we examined the suitability of immunoprecipitation using a His-tagged Prdx2 in the presence of a crosslinker (we chose the DTT-cleavable crosslinker dithiobis-succinimidyl propionate (DSP) [37,38]), which we called “DSP-IP”, and BioID as methods for the identification of Prdx2 interactors induced by H2O2 in mammalian cells. Based on the results of the characterisation of the constructs utilised for both methods to assess the influence of the tags on the properties of Prdx2 and the comparison of the interactor lists obtained with both approaches with other databases, we conclude that BioID is a more reliable method than DSP-IP with His-Prdx2. With BioID we identified 13 proteins that Prdx2 interacts with under elevated H2O2 conditions. Among those, the CSN5 subunit of the COP9 signalosome complex (CSN) warranted further investigation, as CSN5 plays an important role in tumorigenesis and has been shown to interact with STAT3 [39].
In summary, we demonstrate the applicability of BioID for thiol-disulphide unbiased trapping of Prdx interactors in live cells, establish a protocol for its usage, and present new potential interactors of Prdx2. Our results also reveal the importance of Prdx tagged construct characterisation prior to their usage in PPI studies.
2. Materials and methods
2.1. DNA cloning
The expression vector backbone pASIET for BioID constructs was a gift from the lab of Karine Breckpot (VUB) and the genes of BirA*-Flag-Prdx2 and BirA*-Flag-GFP were a gift from the lab of Sebastian Tanco (VIB). The cloning was performed using Gibson assembly. In brief, the pASIET vector was linearised using the BamHI restriction enzyme (Thermo Scientific™ Cat. ER0055) and dephosphorylated by Shrimp Alkaline Phosphatase (Thermo Scientific™ Cat. 783905000UN). The linearised vector was purified by agarose gel electrophoresis using the Wizard® SV Gel and PCR Clean-Up System (Promega Cat. A9281) for extracting the DNA from the gel. The genes of interest were amplified with primers having 20 bp overhangs overlapping the linearised pASIET vector at the 3′ and 5′ ends. The linearised pASIET vector and the overlapping insert genes were assembled using the NEBuilder® HiFi DNA Assembly Master Mix (NEB Cat. E2621) following the manufacturer's protocol and transformed into Stbl3 chemically competent cells (Invitrogen™ Cat. C737303). Positive colonies were selected based on the results of colony PCR. The DNA was prepped using QIAGEN Plasmid Plus Midi Kits (QIAGEN Cat. No./ID: 12943) and confirmed by sequencing. The pCI-His-Prdx2 construct was purchased from the VIB Protein core facility (UGent).
2.2. Cell culture
Human embryonic kidney cell lines (ATCC® CRL-3216™) were cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco™ Cat.11965092) supplemented with 10 % Fetal Calf Serum (Gibco™, Cat.10270106) and 1 % Penicillin-Streptomycin (10,000 U/mL) (Gibco™ Cat.15140122). Cell lines were authenticated by Eurofins and routinely checked for the absence of mycoplasma using the PCR Mycoplasma Test Kit (PromoCell Cat.PK-CA91-1024).
2.3. PEI transfection
Plasmids containing the BioID constructs were introduced into HEK293T by linear Polyethylenimine (PEI 25K, Polysciences Cat.23966-1)-mediated transfection as described previously [40] with minor modifications. Briefly, cells were seeded at 70–80 % confluency in either T75 flasks (CELLSTAR® Cat.658175) or 6-well plates (COSTAR® Cat.3516). Cells were transfected the next day with PEI-complexed plasmids in Opti-MEM™ I Reduced Serum Medium (DNA: PEI ratio 1:2) pre-incubated for 30 min at room temperature. After 4 h of incubation, the medium was replaced with fresh complete growth medium and the cells were allowed to grow overnight.
2.4. Biotinylation of HEK293T cell line
Cells were seeded in 6-well plates and transfected with BioID constructs the next day using PEI as discribed above. After 24 h, the medium was replaced with fresh complete growth medium containing 0 μM, 0.1 μM, 1 μM, 10 μM, 50 μM, or 100 μM biotin. The cells were harvested and lysed for western blot after 24 h of biotinylation.
2.5. CellTiter-Fluor™ Cell viability assay
All experiments were performed in triplicates. Prior to carrying out the cell viability assay using CellTiter-Fluor™ (CTF), a standard curve was built by making a serial dilution of cell numbers in a 96-well plate. HEK293T cells were incubated with 50 μL complete DMEM growth medium and 50 μL 2X CTF solution for 30 min and measured at λex = 380 nm, λem = 505 nm. As a background control, only medium with CTF solution was measured.
2.6. Elevation and quantification of intracellular H2O2 levels
HEK293T cells were seeded at 30 % confluency in a 96-well plate. The next day, the medium was refreshed and the cells were treated with pre-mixed growth medium containing either 1 mU/mL xanthine oxidase dissolved in 50 mM potassium phosphate buffer with increasing amounts of xanthine dissolved in 1 M NaOH (stock concentration 164 mM) (0–10 μM) or increasing concentrations of auranofin dissolved in DMSO (stock concentration 1 mM) (0–0.8 μM), and incubated for 24 h. Then, the medium was discarded, growth medium containing 5 μM of the Peroxy Orange 1 (PO1) H2O2 probe (R&D Systems, Cat. 4944/10) was added to the cells and the cells were incubated at 37 °C for 40 min. Afterwards, the medium was refreshed with 50 μL growth medium and the signal from the probe was measured at λex = 543 nm, λem = 583 nm using a microplate reader (ID5, Molecular Devices). To assess how H2O2 elevation influences cell viability, the CTF assay was used as described above. The normalised PO1 values were calculated using the equation: PO1normalised = PO1sample/(CTFsample-CTFbackground).
2.7. Measuring the HyPer7 probe response in cells
HEK293T cells were transiently transfected with pCI-HyPer7 by PEI transfection (as described above) in a 10 cm TPP® tissue culture dish (Sigma-Aldrich, Cat. Z707686). The next day, transfected cells were trypsinised and re-seeded in 6-well plates. After cells had attached, they were treated with either 8 μM xanthine and 1 mU/mL xanthine oxidase or 0.8 μM auranofin for 24 h. Cells treated with PBS (vehicle) were used as a negative control. The next day, cell images were captured using an Eclipse Ti2 Inverted Microscope (Nikon) with λex = 490 nm, which corresponds to the peak of the HyPer7 excitation spectrum which increases upon oxidation by H2O2 [41]. The area with enhanced fluorescence at λex = 490 nm (%) was calculated by ImageJ using the “analyze→measure” function on 8-bit converted images, which allows the quantification of the area where the signal intensity is in the range 6–60 a.u., where 6 a.u. is the minimum signal value above the background signal and 60 a.u. is the threshold to avoid artificial signals from dead cells. All experiments were performed in triplicates.
2.8. Cell harvesting and lysis
Cells were harvested and lysed following a published protocol with minor modifications [42]. Briefly, cells were washed with PBS and harvested by scraping them off plates with cell scrapers in PBS. The cell suspension was collected and centrifuged at 1,000×g for 5 min. The supernatant was discarded and the weight of cell pellets was determined. The cell pellets were lysed by repeatedly pipetting in complete BioID lysis buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.1 % SDS, 0.2 % Nonidet P-40 substitute, 1.5 mM MgCl2, 1 mM EGTA) containing the cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche Cat.04693132001) or Protease Inhibitor Cocktail (Sigma-Aldrich Cat.P8340) and Benzonase® Nuclease (Millipore Cat.E1014) at a 4:1 v:w ratio. The lysate samples were snap-frozen in liquid nitrogen, and then thawed at 37 °C. The lysate samples were further incubated at 4 °C on an end-over-end rotator for 30 min and centrifuged at 16,000×g for 20 min at 4 °C. The supernatant was then transferred to a new eppendorf tube and the concentration of the proteins in the lysate was determined by a Bradford assay using the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Cat.5000006).
2.9. Western blot analysis
Proteins were separated by SDS-PAGE and transferred onto PVDF membranes (Thermofisher Cat.88518) via the Trans-Blot®Turbo™ System (Bio-Rad). The membranes were blocked in PBS containing 5 % bovine serum albumin (Sigma-Aldrich Cat.A7906) and 1 % Tween20 for 1 h at room temperature. Blots were probed with primary antibodies against beta-actin (mouse host; Sigma-Aldrich Cat.A2228) and biotin using the streptavidin-HRP conjugate (Sigma-Aldrich Cat. 18–152) in PBS containing 3 % bovine serum albumin and 1 % Tween20 at 4 °C overnight. The PVDF membranes were then washed three times with PBS containing 1 % Tween20 and probed with the secondary antibody anti-mouse IgG-HRP conjugate (Invitrogen™ Cat.62-6520) in PBS containing 3 % serum bovine albumin and 1 % Tween20 for 1 h at room temperature. The membranes were again washed three times with PBS 1 % Tween20 and kept in distilled water. Detection of the proteins on the membranes was performed with a chemiluminescence camera using the Pierce™ ECL Western Blotting Substrate (Thermo Scientific™ Cat. 32106). Quantification of signals on the membranes was done by densitometric scanning using the ImageJ software.
2.10. BirA*-Flag-Prdx2 and His-Prdx2 oligomerization assessment
8.8 × 106 HEK293T cells were transfected with pASIET-BirA*-Flag-Prdx2 or pCI-His-Prdx2 using PEI as described above. Cells transfected with pASIET-BirA*-Flag-Prdx2 were treated with 50 μM biotin for 24 h. Cells were treated with DMEM containing either no or 100 μM H2O2 for 15 s. Then the medium was removed and replaced by PBS containing 50 mM NEM for 5 min to block free thiols after which cells were harvested and lysed with Pierce™ IP Lysis Buffer (Thermo Scientific™, Cat.87787), containing protease inhibitors and benzonase. Non-reduced cell lysates and lysates reduced with 50 mM DTT were then analysed by non-reducing SDS-PAGE and native PAGE (Biorad, Cat.4561084) followed by western blot against Prdx2 (mouse host; Sigma-Aldrich Cat. WH0007001M1 or anti-Prdx2 antiserum [43] kindly provided by Bernard Knoops).
2.11. Endogenous Prdx2 immunoprecipitation
50 μL Dynabeads™ Protein G (Invitrogen™, Cat. 10003D) were equilibrated with 50 μL solution of 1 mg/mL BSA in PBS on a rotator for 10 min at 4 °C and then the supernatant was separated using a magnet. 1.5 μg Prdx2 antibody (mouse host; Sigma-Aldrich Cat. WH0007001M1) diluted in 50 μL of a 1 mg/mL solution of BSA in PBS was added to the Dynabeads™ Protein G on a rotator for 1 h at 4 °C. Afterwards the supernatant was separated using a magnet and the Dynabeads™ Protein G were washed twice with 50 μL PBS. The Prdx2 antibody coupled Dynabeads™ Protein G were incubated with untransfected HEK293T cell lysate on a rotator at 4 °C overnight. Then the supernatant was separated using a magnet and the beads were washed 3 times with 200 μL of 10 mM Tris-HCl pH 7.5, 50 mM KCl and eluted in 20 μL of 1 M glycine pH 3. The eluent was quickly neutralised with 2 μL of 1 M Tris pH 10.
2.12. BirA*-Flag-Prdx2 and His-Prdx2 immunoprecipitation
40 × 106 HEK293T cells transfected with pASIET-BirA*-Flag-Prdx2 or pCI-His-Prdx2 using PEI as described above were treated with PBS (vehicle), 0.8 μM auranofin, or 8 μM xanthine and 1 mU/mL xanthine oxidase for 24 h. Cells tranfected with pASIET-BirA*-Flag-Prdx2 were additionally co-treated with 50 μM biotin for 24 h. Cell lysates prepared as above were incubated for 1 h with either Anti-FLAG® M2 Magnetic Beads (Sigma, Cat.M8823) or the Dynabeads™ His-Tag Isolation and Pulldown kit, after which immunoprecipitation with the respective antibodies was performed. The concentration of the pulled down samples was estimated using the Nanodrop by measuring the absorbance at 280 nm and confirmed by SDS-PAGE by loading 20 μL of 6 μM pulled down samples and using purified recombinant Prdx2 as a positive control. The relative band intensity on SDS-PAGE gel was determined densitometrically by ImageJ. For further validation of our pulled down samples they were blotted against Prdx2.
2.13. Peroxidase activity coupled assay
The peroxidase activity coupled assay was conducted following a previously published protocol with minor modifications [[44], [45], [46], [47]]. Buffer exchange into the assay buffer (50 mM sodium phosphate pH 7.4; 150 mM NaCl; 0.1 mM DTPA) was performed for all Prdx2 samples. The human untagged Prdx2 was purchased from Abcam (ab85331). The NADPH oxidation (Δε340 = 6220 M−1cm−1) in the Trx/TrxR/NADPH pathway was measured in function of time at 340 nm in a 96-well plate (Polysorb, Nunc) in a Spectramax 340 PC (Molecular Devices). The reaction mixtures contained 500 μM NADPH, 1 μM TrxR from C. glutamicum (CgTrxR), 8 μM Trx from C. glutamicum (CgTrx), and 0.5 μM recombinant human Prdx2, or 0.5 μM BirA*-Flag-Prdx2, or 0.5 μM His-Prdx2. All reactions were first incubated for 5 min at room temperature and till a stable baseline was observed. The reaction was started by the addition of 20 μM H2O2. Negative control measurements were performed in the absence of H2O2. All data were normalised by subtracting corresponding negative controls as the background value and setting the time point when a decrease in absorbance started to be observed as time point 0. This was necessary to avoid the initial fluctuation in measurements before equilibration of the system. Data were fitted with an exponential decay in Prism9.
2.14. BirA*-Flag-Prdx2 and His-Prdx2 co-immunoprecipitation with STAT3
40 × 106 HEK293T cells were transfected with pASIET-BirA*-Flag-Prdx2 or pCI-His-Prdx2 using PEI as described above. Cells transfected with pASIET-BirA*-Flag-Prdx2 were additionally treated with 50 μM biotin for 24 h. Cells were harvested after 24 h and resuspended in 200 μL DMEM containing either no or 100 μM H2O2 for 2 min. The cells were collected by centrifugation at 1000×g for 2 min and resuspended in 200 μL 50 mM NEM in PBS for 5 min. The cells were subsequently pelleted and lysed in Pierce™ IP Lysis Buffer as described above. Untransfected HEK293T cells were used as a negative control. The cell lysates were then incubated with 50 μL Anti-FLAG® M2 Magnetic Beads or the Dynabeads™ His-Tag Isolation and Pulldown kit on an end-over-end rotator overnight at 4 °C. The next day, the beads were washed 3 times with 500 μL TBS and then the proteins were eluted in the respective solutions according to the product manual. The eluents and whole-cell lysates were analysed by non-reducing western blot using antibodies against STAT3 (Cell Signaling, Cat.9139S). Then the membrane was stripped using the Restore™ PLUS Western Blot Stripping Buffer (Thermo Scientific™, Cat.46430) and re-blotted against Prdx2.
2.15. BirA*-Flag-Prdx2 and CSN5 co-immunoprecipitation
40 × 106 HEK293T cells transfected with pASIET-BirA*-Flag-Prdx2 using PEI as described above were treated with PBS (2 samples), 0.8 μM auranofin, or 8 μM xanthine and 1 mU/mL xanthine oxidase, as well as 50 μM biotin for 24 h. Cells were subsequently harvested by centrifugation at 1,000×g for 5 min and one of the PBS-treated samples was resuspended in 200 μL DMEM containing 100 μM H2O2 for 2 min, after which the cells were pelleted as described. All cell pellets were then resuspended in 200 μL PBS containing 50 mM NEM for 5 min and then collected again. The following procedures were the same as for the BirA*-Flag-Prdx2 and STAT3 co-immunoprecipitation, except that the western blot was performed using antibodies against CSN5 (Cell Signaling Technology, Cat.6895).
2.16. Mass spectrometry sample preparation
Mass spec samples were prepared following a published DiDBiT (Direct Detection of Biotinylated Proteins) protocol [48] with minor modifications. Briefly, cell lysates were precipitated by adding three volumes of methanol, one volume of chloroform, and three volumes of water, vortexed, and centrifuged at 15,000×g for 2 min at room temperature. The supernatant was removed carefully and the precipitated protein layers at the interface were further washed and pelleted by adding three volumes of methanol and centrifuging at 15,000×g for 2 min. Subsequently, the protein pellets were air-dried for 10 min after removing the methanol. The protein pellets were then resuspended in 200 μL of a buffer containing 4 M urea, 50 mM NH4HCO3 and sonicated at 80 % amplitude (20 kHz) for 30 s. The protein suspension was reduced by 5 mM tris(2-carboxyethyl)- phosphine (TCEP) at 55 °C for 20 min and then alkylated by 10 mM iodoacetamide in the dark at 25 °C for 20 min with vigorous shaking using a Thermomixer Eppendorf Comfort at 1000 rpm. Afterwards, the proteins were digested by adding 250 μL 50 mM NH4HCO3, 2.5 μL 1 % ProteaseMAX™ Surfactant (Promega, Cat.V2071) dissolved in 50 mM NH4HCO3, and 1:100 (enzyme/protein, w/w) Sequencing Grade Modified Trypsin (Promega, Cat. V5111) to reach a final reaction volume of 500 μL. The digestion reactions were incubated overnight at 37 °C with vigorous shaking using a Thermomixer Eppendorf Comfort at 1,000 rpm. The next day, the protein digestion reactions were stopped by adding trifluoroacetic acid (TFA) to 0.1 % final concentration. The digested samples were centrifuged at 20,000×g for 20 min at room temperature and the supernatant containing the peptide mixture was transferred to a microcentrifuge tube. Any peptides remaining in the insoluble pellet were extracted by adding 0.5 mL 0.1 % TFA in water, resuspending the pellet by pipetting and centrifuging again for 20 min. The supernatant was pooled together with the previous one and desalted using a Bond Elut C18 EWP cartridge (Agilent, Cat. 12102136). Prior to loading the peptide samples, the cartridges were washed sequentially with 3 mL acetonitrile (ACN), 3 mL 0.5 % acetic acid, 50 % ACN, and 3 mL 0.1 % TFA. After loading the peptide samples, the cartridges were washed with 3 mL 0.1 % TFA and then with 0.250 mL 0.5 % acetic acid in water. The peptides were eluted into an eppendorf tube with 1 mL 0.5 % acetic acid, 80 % ACN in water, and dried in a Speed Vac. Then the dried peptide pellets were re-solubilised in 0.4 mL PBS and the biotinylated peptides were enriched by Pierce™ NeutrAvidin™ Agarose (Thermo Scientific™, Cat. 29200). Prior to incubation, 200 μL slurry of NeutrAvidin resin was loaded to a spin cup (Thermo Scientific™, Cat. 69700) and washed three times with PBS. The solubilised peptide solutions were incubated with NeutrAvidin resin in the same spin cup at 4 °C with rotating overnight. The next day, the spin cups were centrifuged at 1,000×g for 5 min and the flow through were collected as unbound peptides. Resins were washed and centrifuged sequentially with 3 mL PBS, 3 mL 5 % ACN in PBS, and 1 mL ultrapure water. Then biotinylated peptides were eluted by adding 0.3 mL solution containing 0.2 % TFA, 0.1 % formic acid (FA), and 80 % ACN in water. The resins were centrifuged at 1,000×g for 1 min and the eluted biotinylated peptides were transferred to low bind microcentrifuge tube. The second elution of 0.3 mL together with resin in the spin cup were boiled at 95 °C for 5 min for maximum release of peptides from the resin before centrifugation. Finally, the eluted peptides were combined and dried in a Speed Vac and stored at −20 °C until mass spec analysis.
2.17. Co-IP pulldown with and without DSP crosslinker
20 × 106 HEK293T cells transfected with pCI-His-Prdx2 using PEI as described above were treated with PBS (vehicle), 0.8 μM auranofin, or 8 μM xanthine and 1 mU/mL xanthine oxidase for 24 h. Before harvesting the cells, one set of the cells from each condition was washed twice with PBS and incubated with 2 mM syringe-filtered dithiobis(succinimidyl propionate) (DSP) (Thermo Scientific™ Cat. 22585) in PBS for 30 min at room temperature, while the other was left untreated. The crosslinking reaction was then quenched by adding 20 mM Tris/HCl, pH 7.5 for 15 min at room temperature. The cells were lysed by gentle pipetting with 1 mL of RIPA buffer (Thermo Scientific™, Cat.89900) containing the Protease Inhibitor Cocktail (Sigma-Aldrich, Cat.P8340) and Benzonase® Nuclease (Millipore Cat.E1014). The His-Prdx2 was then immunoprecipitated from the cell lysates using the Dynabeads™ His-tag Isolation & Pulldown kit (Invitrogen™, Cat.10103D) following the manufacturer's protocol. The eluted samples were loaded onto an SDS-PAGE gel and transferred to a Turbo™ blot prepacked membrane (BIO-RAD, Cat.1704156). The western blot was incubated as previously described with primary antibodies against Prdx2 (mouse host; Sigma-Aldrich Cat. WH0007001M1), His-tag (mouse host; BIO-RAD Cat.MCA1396), CSN5 (rabbit host; Cell Signaling Technology Cat.6895) and the secondary anti-mouse IgG-HRP conjugate and anti-rabbit IgG-HRP conjugate (Sigma-Aldrich, Cat. GENA934) antibodies.
2.18. Mass spectrometry and statistic analysis
Peptides were dissolved in solvent A (0.1 % TFA in 2 % ACN), directly loaded onto a reversed phase pre-column (Acclaim PepMap 100, Thermo Scientific) and eluted in backflush mode. Peptide separation was performed using a reversed-phase analytical column (Acclaim PepMap RSLC, 0.075 × 250 mm, Thermo Scientific) with a linear gradient of 4%–27.5 % solvent B (0.1 % FA in 98 % ACN) for 100 min, 27.5%–40 % solvent B for 10 min, 40%–95 % solvent B for 1 min and holding at 95 % for the last 10 min at a constant flow rate of 300 nl/min on an Ultimate-3000 RSLC system (ThermoFisher Scientific). The peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) in Fusion Lumos coupled online to the nano-LC. Intact peptides were detected in the Orbitrap at a resolution of 120,000. Peptides were selected for MS/MS using HCD setting at 35; ion fragments were detected in the Ion Trap. A data-dependent procedure that alternated between one MS scan followed by MS/MS scans was applied for 3 s for ions above a threshold ion count of 5 × 103 in the MS survey scan with 30.0 s dynamic exclusion. The electrospray voltage applied was 2.1 kV MS1 spectra were obtained with an AGC target of 4 × 105 ions and a maximum injection time of 50 ms, and MS2 spectra were acquired with an AGC target of 5 × 104 ions and a maximum injection time of 100 ms. For MS scans, the m/z scan range was 350–1500. The resulting MS/MS data was processed using Sequest HT search engine within Proteome Discoverer 2.3 against a human protein database obtained from Uniprot (87,489 entries). Trypsin was specified as cleavage enzyme allowing up to 2 missed cleavages, 4 modifications per peptide and up to 5 charges. Mass error was set to 10 ppm for precursor ions and 0.5 Da for fragment ions. Oxidation on Met, biotinylation on Lys were considered as variable modifications. False discovery rate (FDR) was assessed using Percolator and false positive thresholds for protein, peptide and modification site were specified at 1 %. Abundance ratios were calculated by Label Free Quantification (LFQ) of the precursor intensities within Proteome Discoverer 2.3.
Ratios were calculated as the median of all possible pairwise peptide ratios calculated between replicates using only biotin containing peptides, the maximum allowed fold change was set at 100. The application then used the paired (background-based) method to calculate the p-value and adjusted p-values (Benjamini-Hochberg).
2.19. Mass spectrometry data filtering
Prdx2 interactome candidates in conditions of PBS, auranofin and xanthine/xanthine oxidase from both BioID and DSP crosslinking immunoprecipitation (DSP-IP) were identified from the Proteome Discoverer result file using the following filtering criteria: Abundance ratios: , and , respectively. Abundance Ratio Adj. p-values: , and , respectively. Moreover, for BioID, to be considered a hit, at least one of the identified peptides from the multiple peptides that identify a protein must be biotinylated.
Upregulated Prdx2 interactome candidates upon auranofin treatment from BioID and DSP-IP were identified from the Proteome Discoverer result file using the following filtering criteria: Abundance ratios: and . Abundance Ratio Adj. p-values: and . Moreover, .
Upregulated Prdx2 interactome candidates upon xanthine/xanthine oxidase treatment from BioID and DSP-IP were identified from the Proteome Discoverer result file using the following filtering criteria: Abundance ratios: and . Abundance Ratio Adj. p-values: and . Moreover, .
Downregulated Prdx2 interactome candidates in both conditions from BioID were identified using the same criteria as upregulated ones described above, except Abundance ratios: .
2.20. Venn diagram analysis
For each condition, the overlapping of interactors from BioID and DSP lists were analysed using the online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). The sizes of the circles were then manually adjusted to reflect the number of proteins.
2.21. Proximity ligation assay (PLA)
Prior to seeding, each channel of an μ-Slide VI 0.4 (Ibidi, Cat. 80606) was coated with 30 μL 100 μg/mL Poly-l-lysine (Sigma-Aldrich, Cat.P6282) in PBS for 30 min at room temperature and then washed three times with 100 μL PBS. 35 μL HEK293T suspension (3 × 105 cells/mL) was added per channel gently and incubated for 15 min at 37 °C and 5 % CO2 in the incubator. Then each channel was filled with 120 μL complete DMEM growth medium and the incubation was continued overnight. The next day prior to cell treatment, the old medium was removed and the channels were washed three times with 120 μL PBS (same wash used below). Cells in each channel were treated with 35 μL complete DMEM growth medium containing PBS (vehicle), 0.8 μM auranofin (Sigma-Aldrich, Cat.A6733) or 8 μM xanthine (Sigma-Aldrich, Cat.X0626) and 1 mU/mL xanthine oxidase (Sigma-Aldrich, Cat.X4376) for 24 h. After treatment, the medium was removed and the channels were washed. For cell fixation, the channels were filled twice with 35 μL 4 % formaldehyde in PBS (PFA) and incubated for 20 min at room temperature. For cell permeabilization, the channels were washed and filled twice with 35 μL 0.1 % Triton X-100 in PBS for no more than 3 min. Then, for each sample a proximity ligation assay was performed following the Duolink® PLA Fluorescence Protocol with Duolink™ In Situ PLA® Probe Anti-Rabbit PLUS (Sigma-Aldrich, Cat. DUO92002), Duolink™ In Situ PLA® Probe Anti-Mouse MINUS (Sigma-Aldrich, Cat. DUO92004), Duolink™ In Situ Detection Reagents FarRed (Sigma-Aldrich, Cat. DUO92013), Duolink™ In Situ Wash Buffers, Fluorescence (Sigma-Aldrich, Cat. DUO82049) and Duolink™ In Situ Mounting Medium with DAPI (Sigma-Aldrich, Cat. DUO82040). The primary antibodies of Prdx2 and CSN5 used in this experiment after blocking were diluted at a 1:350 ratio and incubated overnight at 4 °C. Cell fluorescence images were captured using the Eclipse Ti2 Inverted Microscope (Nikon) with a Far-Red filter set.
3. Results
3.1. Tagging Prdx2 with an N-terminal His-tag, but not BirA*, affects Prdx2 peroxidase activity
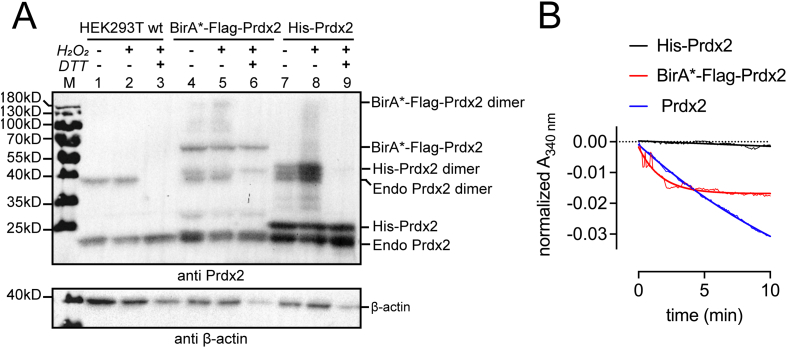
Initially, we attempted to perform DSP-IP with endogenous Prdx2 using Dynabeads coupled to an anti-Prdx2 antibody, as in Pace et al. [49]. However, this approach proved unsuccessful (data not shown) presumably due to insufficiently high Prdx2 levels in HEK293 cells. Indeed, most examples of endogenous Prdx2 purification from mammalian cells use erythrocytes, where Prdx2 levels are known to be particularly high [50,51], or other blood cells, such as Jurkat cells [49]. Hence, we resorted to using an N-terminal His-tagged Prdx2, chosen due to the high affinity of the His-tag to its antibody (KD ~ 10−9 M; while that of a Flag tag is 10−7 M). As tagging the mitochondrial 2-Cys peroxiredoxin Prdx3 has been reported to affect its oligomerization, as well as exert an inhibitory effect on its peroxidase activity [52], we first examined the effect of N-terminal tagging of Prdx2 with either a His-tag, or BirA* (BioID). To this end, we performed several experiments. First, on a native PAGE gel we assessed the oligomeric state of His-Prdx2 (23 kDa) and BirA*-Flag-Prdx2 (60.2 kDa) with and without treatment with a bolus of H2O2. Prdx2 is expected to exist prevalently as decamers under non-oxidative conditions that then disassemble into dimers upon oxidation [53]. As can be seen on Fig. S1A, BirA*-Flag-Prdx2 migrates as a band around 600 kDa, corresponding to the molecular weight of the (homo)decamer, while the band under 480 kDa could be (hetero)decameric endogenous Prdx2. His-Prdx2 decamerises (band around 230 kDa) as well. We could not detect dimer formation upon oxidation, presumably because the bands were too faint to be seen (compare with BirA*-Flag-Prdx2 (lane 5) in Fig. S1A). Next, we asked whether the tagged constructs form reversible disulphide-linked dimers. As can be seen on the non-reducing SDS-PAGE presented on Fig. 1A, both BirA*-Flag-Prdx2 and His-Prdx2 form a dimer that could be reduced by DTT to a monomer.
Fig. 1.
Tagging Prdx2 does not impede reversible dimerisation upon oxidation, but an N-terminal His-tag affects Prdx2 peroxidase activity. (A) HEK293T cells were transfected with BirA*-Flag-Prdx2 or pCI-His-Prdx2 and BirA*-Flag-Prdx2 transfected cells were additionally treated with 50 μM biotin with for 24 h. Cells were treated with or without 100 μM H2O2 for 15 s, followed by 50 mM NEM for 5 min before lysis and then analysed by non-reducing SDS-PAGE with subsequent blotting against Prdx2. The samples in lanes 3, 6, 9 are treated with 50 mM DTT. Beta-actin was used as a loading control. (B) Coupled assay of peroxidase activity. Progress curves for the consumption of NADPH at 340 nm for 0.5 μM pulled down BirA*-Flag-Prdx2, His-Prdx2, and recombinant human Prdx2 in the presence of 500 μM NADPH, 1 μM CgTrxR and 8 μM CgTrx are shown. The reaction was started with the addition of 20 μΜ H2O2. Data were normalised for the reaction in the absence of H2O2 and fitted with an exponential decay in Prism 9.
Finally, we sought to establish whether the BirA*-Flag-tag or the His-tag affect the peroxidase activity of Prdx2. For this purpose, we pulled down the constructs from transfected HEK293 cells, verified their purity (Fig. S1B), and assessed their H2O2-scavenging properties using a coupled peroxidase activity assay. As can be seen in Fig. 1B, BirA*-Flag-Prdx2 scavenges H2O2 with an initial rate comparable to wt (untagged) Prdx2, whereas the activity of His-Prdx2 is severely compromised. While these findings may appear counterintuitive, as a His-tag is often chosen because its small size is thought to be less likely to interfere with protein structure and activity than bigger tags, they are in line with previous studies. As such, the N-terminal His-tagging of Prdx3 exerted an inhibitory effect on its peroxidase activity [52], but it was not affected in the mCER-Prdx2 fusion protein [54].
In summary, our results suggest that while the oligomerization and ability to reversibly form dimers upon the reaction with H2O2 is not substantially affected in either construct, His-Prdx2 has a substantially lower peroxidase activity compared to the BirA*-Flag-Prdx2 construct. We thus proceeded to investigate if and how these differences would translate into the interaction partners we find using both approaches, though these results already warranted caution with interactors found by DSP-IP using the His-Prdx2 construct.
3.2. Application of BioID in HEK293T cells yields biotinylated interactors of Prdx2 and does not interfere with signalling, unlike His-Prdx2
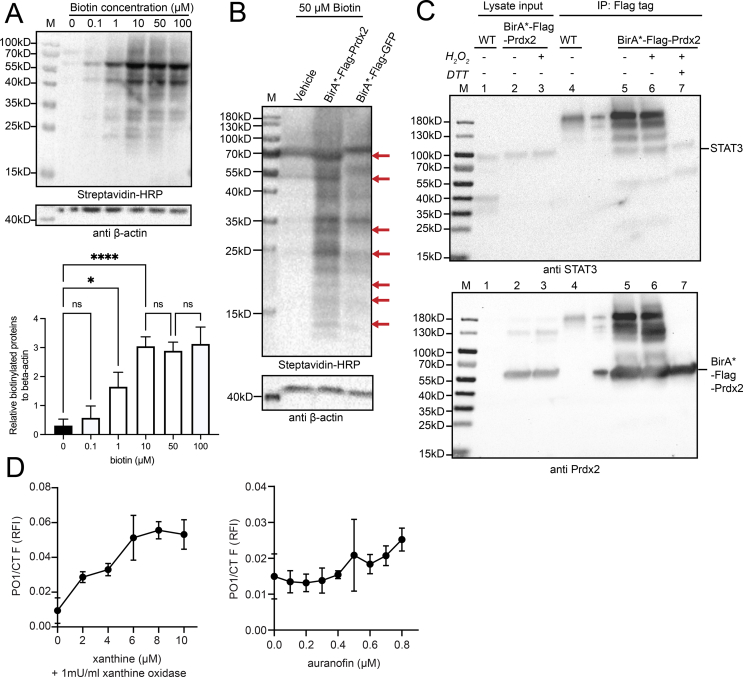
As this is the first application of BioID to detect interactors of Prdx2, or of a redox active protein on the whole, an optimisation of BioID trapping conditions in HEK293T cells was required. First, we established the optimal biotin concentrations needed to biotinylate proteins in the vicinity of Prdx2. HEK293T cells were transiently transfected with a construct encoding BirA*-Flag-Prdx2 and incubated with increasing concentrations of biotin for a period of 24 h. The degree of biotinylation was assessed by western blot (Fig. 2A). As shown on the western blot and on the related densitometric quantification of the blot, background biotinylation was minimal and protein biotinylation reached saturation at 10 μM biotin. However, based on the fact that biotinylation levels were the same for 10 μΜ and 50 μΜ biotin, we opted for the latter as the optimal concentration for all further experiments, as it has been the one used in essentially all previous BioID studies, e.g. Ref. [30]. To verify how specific the biotinylation of Prdx2 as a bait is, we utilised a vehicle and BirA*-Flag-GFP construct (65.3 kDa) as negative controls and checked for differences in the band intensity and pattern between the samples by western blot (Fig. 2B). Under the same experimental conditions, the biotinylation pattern of potential Prdx2 interactors clearly differs from both negative controls with similar expression level of the constructs (Fig. S2C). Therefore, we confirmed that under the selected experimental conditions, BioID can be applied for trapping Prdx2 interactors in HEK293T cells.
Fig. 2.
Application of proximity-dependent biotin labelling in HEK293T allows the capture of proteins specifically interacting with Prdx2, and BirA*-Flag-Prdx2 can co-IP with STAT3 under BioID conditions. Xanthine/xanthine oxidase and auranofin can elevate H2O2 levels in the timeframe needed for the BioID assay. (A) Biotin concentration in the range of 10–100 μM is sufficient to reach maximum biotinylation efficiency. HEK293T cells transiently transfected with the BirA*-Flag-Prdx2 construct were treated with increasing concentrations of biotin ranging from 0 to 100 μM for 24 h and analysed by blotting with streptavidin-horseradish peroxidase (HRP) and anti-beta-actin antibodies (upper panel). Quantification of the biotinylated protein level normalised to beta-actin by ImageJ from the western blot in the lower panel. The graph presents the mean ± SD of data obtained from three independent experiments (Fig. S2A); significance was analysed by a One-Way ANOVA; n.s., not significant; *p < 0.05; **p < 0.01 (lower panel). (B) BioID biotinylation for the Prdx2 bait protein differs from the negative GFP control. HEK293T cells transiently transfected with vehicle, BirA*-Flag-Prdx2, or BirA*-Flag-GFP constructs were incubated for 24 h with 50 μM biotin and analysed by blotting with streptavidin-HRP and anti-beta-actin antibodies. Red arrows highlight the distinctly different biotinylated protein patterns between the BirA*-Flag-Prdx2 and BirA*-Flag-GFP samples. The figure represents a single western blot from three independent experiments (Fig. S2A). (C) BirA*-Flag-Prdx2 can co-IP with STAT3 under BioID conditions. BirA*-Flag-Prdx2 transfected cells were supplemented with 50 μΜ biotin for 24 h, harvested and subsequently exposed to either 0 μM or 100 μM H2O2 for 2 min followed by 50 mM NEM for 5 min. Untransfected HEK293T cells were used as negative control (wt). Samples were blotted against STAT3 (upper panel), and after stripping re-blotted against Prdx2 (lower panel). (D) Prolonged induction of intracellular H2O2 for 24 h in HEK293T with (left panel) 1 mU/ml xanthine oxidase with xanthine concentrations ranging from 0 to 10 μM, and (right panel). Auranofin concentrations ranging from 0 to 0.8 μM H2O2 levels were evaluated with the Peroxy Orange 1 probe (PO1, λex = 543 nm, λem = 583 nm) and normalised to the number of viable cells determined using the CellTiter-Fluor™ kit (CTF, λex = 380 nm, λem = 505 nm) as relative fluorescence intensity (RFI). 8 μM xanthine/1 mU/ml xanthine oxidase and 0.8 μM auranofin were selected as optimal conditions for prolonged intracellular H2O2 induction, which was confirmed with the HyPer7 probe in vivo (Fig. S2D). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To ensure that BioID conditions of 24 h treatment with 50 μΜ biotin and fusion of BirA*-Flag to Prdx2 do not interfere with Prdx2 signalling, i.e. from interacting with its known partners, we tested whether we could pull down STAT3, a well-established interactor of Prdx2 [15]. As can be seen in Fig. 2C, we could co-immunoprecipitate STAT3 with BirA*-Flag-Prdx2 following a 24 h treatment with 50 μΜ biotin. This result also shows that the expression of BirA*-Flag-Prdx2 per se also does not disrupt signalling. Of note, parallel experiments aiming to co-immunoprecipitate STAT3 with the His-Prdx2 construct did not yield any results (data not shown), forming yet another indication of this construct being suboptimal for studying Prdx2 interactors.
As in this study we specifically aim to find H2O2-mediated Prdx2 interactors, we sought to establish a system that would lead to elevated H2O2 levels in HEK293T cells over the 24 h required for maximum biotinylation [55]. We decided to test the xanthine/xanthine oxidase (X/XO) H2O2 generating system [56,57], and auranofin, a TrxR inhibitor [58] which blocks the electron transfer from NADPH to thioredoxin, and as a consequence to all the thioredoxin-dependent peroxidases, including Prdx2. Since reduced Prdx2 is one of the most efficient and abundant intracellular H2O2 scavengers [59], this is expected to lead to an increase in intracellular H2O2 levels. These two systems thus differ in the site of H2O2 generation: while xanthine/xanthine oxidase generates H2O2 exogenously, which then enters the cell via aquaporins [60], auranofin leads to an intracellular build-up of H2O2. To this end, HEK293T cells were treated with increasing concentrations of X/XO (Fig. 2D) and auranofin (Fig. 2E) for 24 h after which the H2O2 levels were assessed using the Peroxy Orange 1 (PO1) H2O2 probe, which we normalised to the number of live cells as detected using the CellTiter-Fluor™ Cell Viability Assay (CTF) carried out in the same cells to account for cell death due to treatment. As can be seen in Fig. 2D, the PO1/CTF ratio gradually increased with increasing xanthine concentrations and reached a plateau at 8 μM xanthine, which we selected for further experiments. As for auranofin, we chose 0.8 μΜ, as at this concentration we observe an increase in H2O2 levels and it is in agreement with the concentrations typically used in the redox biology literature, e.g. Ref. [61]. To confirm that these concentrations of X/XO and auranofin increase the H2O2 levels, we used an alternative readout in the form of the ultrasensitive and pH-independent HyPer7 probe [41]. For this purpose, we transiently transfected HEK293T cells with the cytosolic HyPer7, treated them either with the vehicle (PBS), X/XO, or auranofin (Fig. S2D), and evaluated the fluorescence intensity of the probe under the microscope at λex = 490 nm, which corresponds to the HyPer7 wavelength at which the intensity increases upon oxidation. Quantification of the fluorescence intensity (Fig. S2D) indicated that treatment with X/XO and auranofin increased the signal intensity by approximately two-fold. Therefore, we confirmed that the selected concentrations of xanthine (8 μM)/xanthine oxidase (1 mU/mL), and 0.8 μM auranofin are inducing intracellular H2O2 production after 24 h, required for applying the BioID approach.
3.3. BioID and DSP-IP yield different sets of Prdx2 interactors
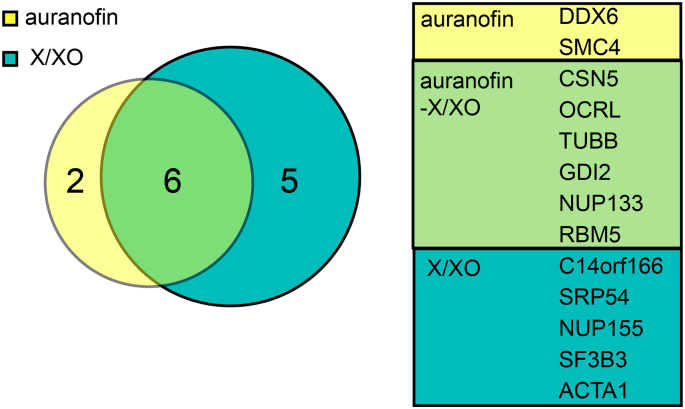
We next proceeded to identify the Prdx2 interactors obtained by BioID and DSP-IP by mass spectrometry upon an elevation of H2O2 levels. For the BioID mass spectrometric approach, cells were transfected with expression constructs for BirA*-Prdx2 and BirA*-GFP, and after 24 h, treated with 50 μM biotin for another 24 h. Intracellular H2O2 levels were increased by applying the selected concentrations of auranofin and X/XO in culture for 24 h at the same time as biotin. Lysates were enriched for biotinylated peptides and identified by mass spectrometry. To exclude possible false positive interactors, the list of the Prdx2 interactome was stringently filtered. To be considered a Prdx2 interactor under elevated H2O2 conditions, a protein hit had to be abundantly found in either the auranofin or X/XO-treated sample. “Abundant” proteins are those that have an abundance ratio: (auranofin treatment)/(negative control) score 100 and a value greater than 1 in the abundance ratio: (auranofin treatment)/(PBS treatment) or an abundance ratio: (xanthine/xanthine oxidase treatment)/(negative control) score 100 and a value greater than 1 in the abundance ratio: (xanthine/xanthine oxidase treatment)/(PBS treatment), respectively. In general, the higher this value, the more significant the increase in the abundance is. The value of 100 is the maximum possible fold value set arbitrarily by the Proteome Discoverer 2.3 software. In addition, the posterior error probability (Sum PEP score), i.e. the likelihood that the peptide spectrum match is correct, had to be The application of these selection criteria resulted in the thirteen candidates presented in Table 1, while the detailed mass spec data is provided in the Excel sheet, “ox BioID versus PBS" tab. Notably, six candidates (marked with asterisks in Table 1): TUBB, GDI2, NUP133, RBM5, OCRL and CSN5 were upregulated in both X/XO and auranofin treatment conditions, i.e. upon both exogenous and endogenous H2O2 generation (Fig. 3). Several downregulated candidates in both conditions were also identified and are listed in Table S1. In addition, all interactors that were identified by BioID, including in the PBS-treated samples, following filtering, can be found in the supplementary excel sheet “BioID BirA*-Prdx2 interactors” tab, whereas unfiltered data is presented in the “raw data” tab.
Table 1.
List of potential Prdx2 interactors from BioID after data filtering for upregulated Prdx2 interactome with indication of accession, description, and values in abundance ratio: (auranofin treatment)/(negative control); abundance ratio: (xanthine/xanthine oxidase treatment)/(negative control); abundance ratio: (auranofin treatment)/(PBS treatment) and abundance ratio: (xanthine/xanthine oxidase treatment)/(PBS treatment). And additional information of numbers of cysteine residues and cited sulphenylation sites.
| UniProt Accession |
Description and Gene name | Abundance Ratio: (auranofin treatment)/(negative control) | Abundance Ratio: (xanthine/xanthine oxidase treatment)/(negative control) | Abundance Ratio: (auranofin treatment)/(PBS treatment) | Abundance Ratio: (xanthine/xanthine oxidase treatment)/(PBS treatment) | Numbers of Cysteine residues | Sulphenylation sites |
|---|---|---|---|---|---|---|---|
| P07437a | Tubulin beta chain TUBB |
100 | 100 | 100 | 100 | 8 | |
| P50395a | Rab GDP dissociation inhibitor GDI2 |
100 | 100 | 100 | 100 | 8 | C277, C302 [62] |
| Q8WUM0a | Nuclear pore complex protein Nup133 NUP133 |
100 | 100 | 100 | 100 | 18 | |
| P26196 | Probable ATP-dependent RNA helicase DDX6 DDX6 |
100 | 100 | 7 | C102 [62] | ||
| P52756a | RNA-binding protein 5 RBM5 | 100 | 100 | 100 | 100 | 9 | |
| Q01968a | Inositol polyphosphate 5-phosphatase OCRL-1 OCRL |
100 | 100 | 100 | 100 | 21 | |
| Q9NTJ3 | Structural maintenance of chromosomes protein 4 SMC4 | 100 | 100 | 0.01 | 12 | C916 [62] | |
| Q92905a | COP9 signalosome complex subunit 5 JAB1/CSN5/COPS5 |
100 | 100 | 9.446 | 13.886 | 4 | C218 [62] |
| P68133 | Actin, alpha skeletal muscle ACTA1 | 19.794 | 100 | 1 | 100 | 6 | |
| Q15393 | Splicing factor 3B subunit 3 SF3B3 | 15.795 | 100 | 8.318 | 100 | 19 | C269, C1054, C1156 [62] |
| P61011 | Signal recognition particle 54 kDa protein SRP54 |
0.755 | 100 | 1 | 100 | 5 | |
| Q9Y224 | RNA transcription, translation and transport factor protein C14orf166 |
100 | 0.01 | 100 | 2 | ||
| O75694 | Nuclear pore complex protein Nup155 NUP155 |
1 | 100 | 0.869 | 100 | 28 |
Six potential Prdx2 interactor candidates marked with an asterisk are upregulated in both H2O2 elevating conditions.
Fig. 3.
Venn diagram depicting the number of shared upregulated Prdx2 interactors between auranofin and xanthine/xanthine oxidase (X/XO) identified by BioID. The diameter of the circles reflects the number of proteins in each one. More details are listed in Table 1.
Next, we performed DSP-IP on cells that were also treated with the same concentrations of X/XO or auranofin for 24 h. Application of the same criteria as for BioID on the list of proteins obtained by mass spec yielded a list of nineteen interacting partners of Prdx2 in the presence of auranofin and xanthine/xanthine oxidase: SPTAN1, U2SURP, ACTBL2, ACTN4, RRP9, CASP14, LIMA1, ANXA1, PINX1, DDX18, GSN, TMOD2, BPIFA1, ASPRV1, TGM3, ATXN2L, ARPC5L, NHP2L1 and CTSD (Table 2). Interestingly, there was absolutely no overlap between the interactors detected upon elevated H2O2 conditions by the two approaches.
Table 2.
List of potential Prdx2 interactors from DSP-IP after data filtering for upregulated Prdx2 interactome with indication of accession, description, and values in abundance ratio: (auranofin treatment)/(negative control); abundance ratio: (xanthine/xanthine oxidase treatment)/(negative control); abundance ratio: (auranofin treatment)/(PBS treatment) and abundance ratio: (xanthine/xanthine oxidase treatment)/(PBS treatment).
| UniProt Accession |
Description and Gene name | Abundance Ratio: (auranofin treatment)/(negative control) | Abundance Ratio: (xanthine/xanthine oxidase treatment)/(negative control) | Abundance Ratio: (auranofin treatment)/(PBS treatment) | Abundance Ratio: (xanthine/xanthine oxidase treatment)/(PBS treatment) |
|---|---|---|---|---|---|
| Q13813-3a | Isoform 3 of Spectrin alpha chain, non-erythrocytic 1 SPTAN1 | 100 | 100 | 2.063 | 100 |
| H3BSK9 | Ataxin-2-like protein (Fragment) ATXN2L |
100 | 0.01 | 100 | 0.01 |
| Q562R1a | Beta-actin-like protein 2 ACTBL2 | 100 | 100 | 3.384 | 100 |
| K7EJH8a | Alpha-actinin-4 (Fragment) ACTN4 | 100 | 100 | 2.602 | 2.855 |
| O43818a | U3 small nucleolar RNA-interacting protein 2 RRP9 |
100 | 100 | 2.36 | 6.889 |
| Q9BPX5 | Actin-related protein 2/3 complex subunit 5-like protein ARPC5L |
100 | 100 | 1.779 | 1.553 |
| F8WBT5a | PIN2/TERF1-interacting telomerase inhibitor 1 PINX1 | 100 | 100 | 100 | 100 |
| Q9NVP1a | ATP-dependent RNA helicase DDX18 DDX18 |
100 | 100 | 100 | 100 |
| P55769 | NHP2-like protein 1 NHP2L1 | 100 | 1 | 100 | 3.302 |
| C9JH19 | Cathepsin D light chain (Fragment) CTSD |
100 | 100 | 100 | 1 |
| O15042 | U2 snRNP-associated SURP motif-containing protein U2SURP | 100 | 100 | ||
| P31944 | Caspase-14 CASP14 |
100 | 0.01 | 3.616 | |
| F8VS07 | LIM domain and actin-binding protein 1 LIMA1 |
100 | 100 | 2.921 | |
| P04083 | Annexin A1 ANXA1 |
100 | 100 | ||
| P06396 | Gelsolin GSN |
100 | 100 | 100 | |
| Q9NZR1 | Tropomodulin-2 TMOD2 |
100 | 100 | ||
| Q9NP55 | BPI fold-containing family A member 1 BPIFA1 |
100 | 100 | ||
| Q53RT3 | Retroviral-like aspartic protease 1 ASPRV1 |
100 | 100 | ||
| Q08188 | Protein-glutamine gamma-glutamyltransferase E TGM3 | 0.01 | 100 | 100 |
Six potential Prdx2 interactor candidates marked with an asterisk are upregulated in both H2O2 elevating conditions.
We next asked how the interactors identified by BioID and DSP-IP would compare to known potential Prdx2 interactors found in the BioGRID4.3 database [63]. Out of all interactors identified in this study by both approaches, only CSN5 (identified by BioID) was present in the database [64].
Just as the techniques used to identify most of the entries in the BioGRID4.3 database, both DSP-IP and BioID are high-throughput approaches, and their results should be validated by low-throughput methods, as it has been estimated that only 30–50 % PPIs identified by high-throughput methods are biologically relevant [65]. Combining the results of the construct characterisation (Fig. 1) with the BioGRID4.3 overlap, we decided that only the BioID results warrant further validation: unlike the His-Prdx2, BirA*-Flag-Prdx2 does not substantially differ in terms of signalling from untagged Prdx2 and displays peroxidase activity (Fig. 1). Of the 13 BioID interactors identified under elevated H2O2 conditions filtered with stringent conditions, we selected CSN5 for further validation. We based our choice on the fact that it was the only protein to have been previously identified as a potential Prdx2 interactor according to the BioGRID4.3 database using other highthroughput methods, in this case, the yeast two-hydrid technology [64], and as it was interacting with Prdx2 independent of the H2O2 source. We also considered the physiological role of CSN5 and its involvement in cancer [66].
3.4. Prdx2 interacts with CSN5 in HEK293T cells
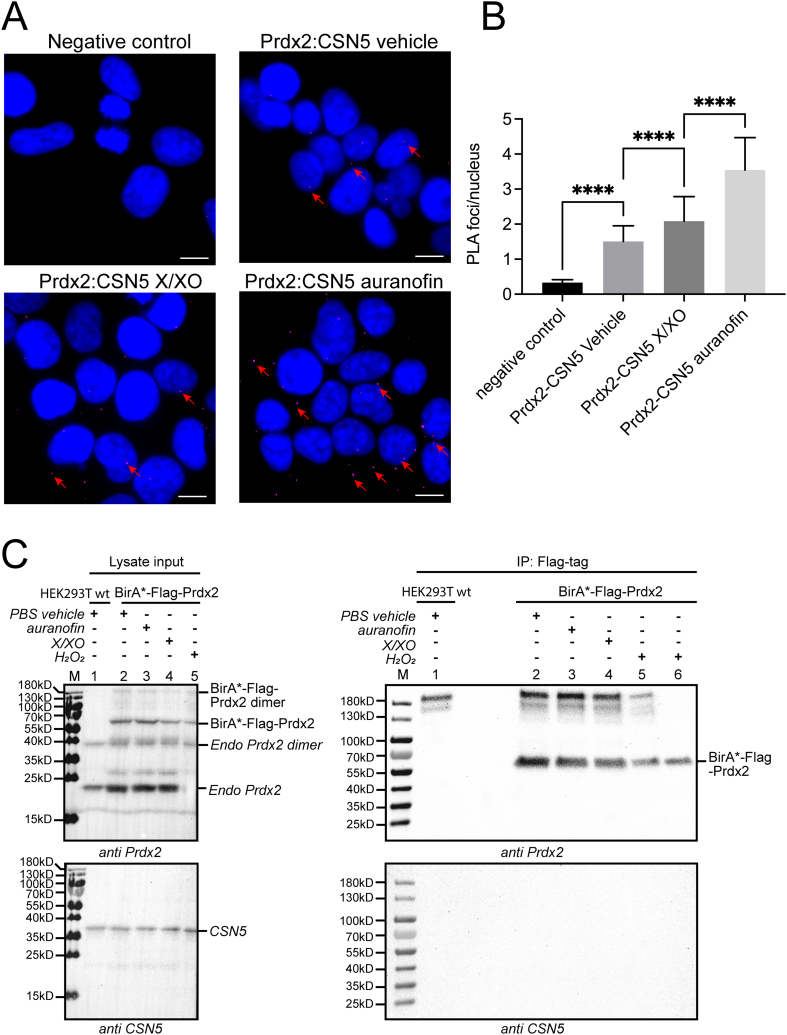
To validate the BioID approach and to provide proof of the Prdx2:CSN5 interaction in a low throughput setting, which had never been done before, we employed two methodologies: the proximity ligation assay (PLA) and co-immunoprecipitation (co-IP). PLA has the additional advantages of enabling the visualisation of PPIs between endogenous proteins, as well as labelling interacting proteins at the level of single molecules. Therefore, it serves as an especially powerful validation method for PPIs detected by techniques requiring transfection, as it ensures that the PPI was not caused by unphysiologically high protein expression levels. Samples incubated with no primary, but only secondary, antibodies, were used as a negative control (Fig. 4A). HEK293T wt cells were stimulated with X/XO or auranofin with the same concentrations and treatment duration as in the BioID experiment. We found that under vehicle and X/XO conditions, the PLA foci per nucleus of Prdx2:CSN5 interactions show a slight increase compared to the negative control. Treatment with 0.8 μM auranofin increases the number of PLA foci per nucleus of Prdx2:CSN5 even further (Fig. 4B). Conversely, in a co-immunoprecipitation experiment using HEK293T cells transfected with constructs expressing BirA*-Flag-Prdx2 and treated with PBS vehicle, auranofin, or X/XO we found no CSN5 to be co-immunoprecipitating, even though CSN5 could be detected in the input (Fig. 4C). As expected from the DSP-IP results, His-Prdx also could not pull down CSN5, even though again endogenous CSN5 could be detected (Fig. S3). Our results are in line with another study that also failed to detect a Prdx2:CSN5 interaction by co-IP, even though, likewise, the expression level of the Flag-tagged constructs was high enough to be detected by western blot [64]. Taken together, these results indicate that endogenous Prdx2 and CSN5 do indeed interact upon an increase in H2O2 levels, yet this interaction is likely either weak, indirect, or transient.
Fig. 4.
The CSN5:Prdx2 interaction upon H2O2 level elevation was confirmed by proximity ligation, but not with co-immunoprecipitation (A) Proximity ligation assay (PLA): HEK293T cells were treated as follows: vehicle – untreated cells; X/XO - cells treated with 8 μM xanthine plus 1 mU/mL xanthine oxidase for 24 h; auranofin - 0.8 μM auranofin for 24 h. The primary antibodies of Prdx2 and CSN5 were used at a 1:350 dilution. The presented images are representative of three biological independent experiments conducted in HEK293T wt cells. The PLA foci are shown in magenta, with some emphasised by red arrows. The DAPI-stained cellular nuclei are in blue. Scale bar: 10 μm. (B) The bar graphs quantify the number of PLA foci per nucleus by ImageJ software above a threshold of 1,500, representing the mean ± SD of data obtained from three independent experiments with overall n = 332 to 379 cell nuclei in each condition, respectively; the significance was calculated using a One-Way ANOVA; ****p < 0.0001 to every other columns. (C) HEK293T cells were transfected with BirA*-Flag-Prdx2, while non-transfected wt cells served as a negative control, and treated with PBS vehicle, 0.8 μM auranofin or 8 μM xanthine plus 1 mU/mL xanthine oxidase, as well as 50 μM biotin for 24 h. Cells were harvested, and PBS vehicle treated cells were resuspended in DMEM with or without 100 μM H2O2 for 2 min. Subsequently, all cells were resuspended in PBS containing 50 mM NEM for 5 min before lysis. Cell lysates were immunoprecipitated (IP) with Anti-FLAG® M2 Magnetic Beads and analysed by non-reducing SDS-PAGE followed by a western blot using anti-Prd×2 antibody and then stripped to reblot using anti-CSN5 antibodies. Sample in lane 6 is the sample of lane 5 reduced with 50 mM DTT. No CSN5 co-immunoprecipitated with BirA*-Flag-Prdx2. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this explorative study, we set the scope of testing the applicability of two thiol-disulphide independent approaches, a proximity labelling method, BioID, and co-immunoprecipitation with the chemical crosslinker DSP (DSP-IP), for the identification of H2O2-induced interactors of Prdx2.
The two approaches yielded lists of interactors with no overlap. The lack of overlap, however, is not unprecedented. For example, Lambert et al., who compared BioID and IP in a study of chromatin-associated complexes [67], also found largely distinct interactomes. On a more general scale, little overlap among different high-throughput approaches has long been observed [68,69]. This may or may not have to do with the stringency of mass spec data filtration, which is often not given sufficient attention (see supplementary excel sheet for all data and data filtering stringency).
There could be several reasons for the discrepancies we observe between BioID and DSP-IP. First, our data (Fig. 1) showed that while the oligomerization and ability to form reversible disulphide-linked dimers upon the reaction with H2O2 is not seemingly affected in constructs used in either approach, the peroxidase activity of His-Prdx2, but not of the BirA*-Flag-Prdx2, is greatly diminished. Prdx2 peroxidase activity is also known to be inhibited by post-translational modifications (PTMs), such as phosphorylation, S-nitrosylation, and glutathionylation [70]. Our observation that differences in peroxidase activity could be responsible for different interactors thus raises an interesting question on the role of these PTMs in mediating Prdx2 partner choice. In general, lamentably few studies that use tagged Prdx2 to study PPIs include a peroxidase activity test, most of them limiting themselves to monitoring the construct size on an SDS-PAGE gel. Such is the case, for example, in studies that employed C-terminal His-tagged [71] and HA-tagged [72], or N-terminal Flag-tagged Prdx2 [18,73]. As most studies included in the BioGRID4.3 database detected Prdx2 interactors with tagged proteins by either yeast two-hybrid or co-IP approaches, this could potentially explain the poor overlap between interactors we report here and those in the database. Though, to our knowledge, it has never been shown, in principle, that tagging could also potentially alter the targeting of Prdx2 to microdomains or even compartments, inevitably leading to different interactors.
Another possible explanation for this result could be the different labelling distances between the two methodologies: 15 nm for BioID [31] and 1.2 nm for DSP [74]. It would be interesting to see if and how longer and more flexible linkers between BirA* and Prdx2 and cross-linkers such as the zero-length cross-linker EDC would affect the proteins identified. The low numbers of interactors identified in our study with stringent mass spec data filtering criteria do not allow us to perform a functional annotation enrichment analysis and classify the proteins according to their biological process GO terms. This could have given us an idea of whether there is a bias of the two approaches towards proteins belonging to a biological process. For example, we would expect that the bigger labelling distance of BioID would allow the trapping of more transient interactors. Indeed, a similar study by Lambert et al. to which we already referred above, reported that BioID and IP identify interactors that belong to different processes [67]. Moreover, the same study found that BioID can trap a larger number of proteins, including those that are less abundant. This observation can in principle also be explained by the longer duration of the BioID experiment (24 h) compared to DSP-IP (30 min). Important to note, we confirmed that the interactors identified by BioID are not merely artefacts of a 24 h treatment with biotin, by showing that the Prdx2:STAT3 interaction still occurs, suggesting that Prdx2 redox signalling is not significantly affected (Fig. 2C). Of note, BirA variants that require times as short as 10 min for biotinylation (TurboID [75]), have been developed, which would perhaps be even more suitable to capture transient redox interactions, but at present they are plagued by instability of the BirA variant, unspecific biotinylation and cell toxicity [76].
Regardless of the precise reasons for the differences observed between the Prdx2 interactors identified by BioID vs DSP-IP, the primary scope of this study was to find a reliable thiol-disulphide independent approach for the identification of Prdx2 interactors. The results of our in-depth comparison of BioID and DSP-IP point in favour of the former: tagging Prdx2 with BirA* does not seem to affect its peroxidase activity and the construct can still pull down STAT3, an established partner of Prdx2.
As a side note, it is curious that neither STAT3, nor AnnexinA2 were identified by either of our two approaches (despite STAT3 being pulled down by the BirA*-Flag-Prdx2 construct). This could be due to the inaccessibility of STAT3 lysine residues to BirA* for biotinylation and to DSP for cross-linking. Indeed, the solved structure of STAT3 (PDB ID: 6QHD) revealed that STAT3 lysine residues are acetylated [77]. Confirming the low overlap of high-throughput PPI studies [68,69], STAT3 is also not present in the BioGRID4.3 database, even though STAT3 could be pulled down in high-throughput studies employing an N-terminal Flag-tagged Prdx2 construct [18], as well as a C-terminal SBP-tagged one [15]. A plausible explanation for the absence of AnxA2 in our interactor lists could be due to the fact that the labelling distance of both BioID and DSP-IP is too small - Talwar et al. investigated the Prdx2:AnxA2 interaction by PLA, which can detect proteins that are as far as 80 nm apart [78], i.e. 55 nm and 78.8 nm more than BioID and DSP-IP, respectively.
To find out whether BioID conforms to the criteria expected from an approach to identify thiol-disulphide independent Prdx2 interactors, we should consider the proteins we identified as Prdx2 interactors in our experiments from the perspective of the mechanism by which they would likely interact with Prdx2. In other words, would these Prdx2 interactors have been found with thiol-disulphide dependent approaches? In total, with BioID we discovered thirteen interacting partners of Prdx2 in the presence of auranofin and xanthine/xanthine oxidase: TUBB, GDI2, NUP133, DDX6, RBM5, OCRL, SMC4, CSN5, ACTA1, SF3B3, SRP54, C14orf166 and NUP155, of which only CSN5 was reported as to be a Prdx2 interactor, and that in a high-throughput setting by yeast two-hybrid technology [64]. To filter the mass spec hits, we compared the hits obtained when using Prdx2 as a “bait” to those with GFP and set the threshold for the abundance ratios of H2O2 treated samples vs PBS (Table 1). Indeed, perhaps the biggest potential peril of the BioID approach lies in selecting the correct negative control and setting the threshold in an unbiased way. Further, as BioID requires the transfection of cells, its applicability in primary and other cells not amenable to transfection is limited. This limitation can turn into an advantage in some settings though, for example, when investigating Prdx2 interactors in cells that have low levels of Prdx2, e.g. HL-60 (Cell Atlas). We could not pull down endogenous Prdx2 in this study in HEK293T cells, and a recent study hinted that Prdx partners and oxidation mechanisms may be cell specific [49]. Furthermore, by attaching specific targeting sequences to the ectopically expressed Prdx2 construct, it is possible to target the BirA*-Flag-tagged bait to different organelles to identify compartment-specific interactors, as has recently been done for mitochondria and other organelles [79,80].
Even though all of the thirteen hits harbour at least two cysteine residues, only GDI2, DDX6, SMC4, SF3B3, and CSN5 were identified to be oxidised to sulphenic acid in a proteome-wide screen with chemical probes following treatment with 0.5 mM H2O2 [62]. It is likely that the same cysteine residues would be the ones forming mixed disulphides with Prdx2, as the same redox-sensitive cysteines are expected to perform a nucleophilic attack on both H2O2, as well as on the oxidised CysP of Prdx2 (whether sulphenylated or disulphide-bonded with CysR) [81].
A vast number of Prdx2 targets seem to form disulphide bonds [14]. For this to happen, apart from the thiol which forms a sulphenic acid, there should be another thiol capable of performing a nucleophilic attack on the sulphenylated cysteine (or, in case of redox relay with Prdx2, on the cysteine forming a mixed disulphide with Prdx2). Of the proteins found to be sulphenylated, crystal structures are available for SMC4 (PDB ID: 4U4P), DDX6 (PDB ID: 4CT5 [82]), SF3B3 (cryo-EM structure PDB ID: 5ZYA [83]) and CSN5 (PDB ID: 5M5Q [84]), and not all include a cysteine found to form a sulphenic acid. Of the ones that do, only in CSN5 could the sulphenylated Cys218 form a disulphide bond (with Cys145), and that only after considerable structural changes, as both cysteines are 15.6 Å apart. However, these cysteines are conserved, and in the Archaeoglobus fulgidus homologue of CSN5, they were reported to form a disulphide bond [85]. Finally, CSN5 was identified as a Trx1 interactor using a thioredoxin trapping mutant and Cys218 was shown to be necessary for the interaction [86]. In any case, the unravelling of the mechanistic details of how CSN5, or the other hits, interact with Prdx2, is beyond the scope of this study. All in all, these considerations suggest that the 8 out of 13 interactors not found to be sulphenylated could have been missed by thiol-disulphide dependent approaches.
We next sought to validate the Prdx2:CSN5 interaction using two other mechanism-independent approaches: co-immunoprecipitation (co-IP) and the proximity ligation assay (PLA). In the case of the former (Fig. 4C), we could not detect a band corresponding to CSN5 following a pull-down with antibodies against tagged Prdx2 even in the presence of DSP (Supplementary Fig. S3). In contrast, the interaction between Prdx2 and CSN5 was confirmed using PLA (Fig. 4A). One plausible explanation is purely mechanistic - as DSP targets primary amines (i.e. Lys residues) in both partners, if no such couple exists at the site(s) of interaction(s), the crosslink will not be made or result in a mono-link and the MS post-acquisition software will not pinpoint the modified residue. Another explanation is that PLA allows the interacting proteins to be further away than the crosslinker: proteins will be linked up to a distance of 80 nm [78], which increases the probability of capturing dynamically interacting proteins, that, for example, remain confined within a microdomain of the cells, such as the membrane, even when not directly interacting. Based on our results, the Prdx2:CSN5 interaction could be dynamic, transient, and/or indirect. In fact, the ability to capture transient and low affinity interactions is hailed as one of the advantages of BioID over IP [87]. Just as in the case of Prdx2:STAT3, it is possible that another protein is needed to scaffold the Prdx2:CSN5 interaction. This could be addressed in follow-up studies by a derivative of BioID termed split-BioID, where the biotin ligase is split between two interaction partners and biotinylation only occurs when the partners come together to allow complementation of the two parts of the biotin ligase [88]. It remains to be seen if other identified BioID interactors can be confirmed in low-throughput studies.
It is also worthy to note that we observed a clear difference between external H2O2 induction via the X/XO system compared with internal H2O2 induction via the inhibition of thioredoxin reductase with auranofin, for both upregulated and downregulated Prdx2 interactors, which cannot be explained by differences in H2O2 as measured with both probes (Fig. 2D and Supplementary Fig. 2). As mentioned above, auranofin increases H2O2 levels by preventing peroxiredoxin recycling by the thioredoxin system. However, the effect of oxidised Prdx2 accumulation caused by auranofin is only observed within the first 2 h of a 24 h course [89,90], and might be compensated by electron transfer via the glutathione reductase system [90]. Indeed, as mentioned above, the glutathionylation of the peroxidatic cysteine of Prdx2 after peroxide challenge has been reported [19]. Of note, as studies investigating Prdx2 interactors typically employ bolus treatments of H2O2, such as 100 μM for 15 s [14], this could be a further explanation for the low overlap of the interactors we find here by BioID and those present in the BioGRID4.3 database. Whether the duration and source of H2O2 alters the mechanism of Prdx2 mediation of protein oxidation is an interesting topic for future investigations.
Regarding the potential biological significance of the Prdx2:CSN5 interaction, CSN5 is the catalytic 37.5 kDa Zn2+-binding subunit of the multisubunit COP9 (Constitutive photomorphogenesis 9) signalling complex (CSN – 320 kDa) with isopeptidase and deNEDDylase activity. Numerous studies have reported that CSN5 is overexpressed in several cancers and is often associated with poor prognosis including breast cancer, nasopharyngeal cancer, lung cancer, and liver cancer [66]. This CSN complex is highly evolutionary conserved and controls 20 % of the cellular ubiquitylation events, thereby influencing almost every pathway [91]. Intriguingly, one of the signalling pathways CSN5 is involved in is STAT3 [66], which has been shown to enhance the transcription of COPS5, the gene of CSN5 [92]. However, as Prdx2-mediated STAT3 oxidation attenuates STAT3 transcriptional activity [15], this effect of STAT3 on CSN5 expression is unlikely to be a direct consequence of Prdx2 oxidation. On the other hand, the NEDDylation of Cullin1, a substrate of the yeast COP, was recently found to be redox-regulated, leading to the speculation that COP activity could also be redox-regulated [93]. The fact that CSN5 was reported to be overexpressed with thioredoxin in patient samples of relapsed acute monocytic leukemia (AML) under oxidative stress [94] further speaks in favour of CSN5 being redox-regulated. If this is the case, it will be interesting to see whether Prdx2 is involved in NEDDylation.
Prdx upregulation has been documented in several cancers [95], yet we are still a long way from a detailed understanding of Prdx-mediated redox signalling in cancer. An elucidation of the interacting partners with both thiol disulphide-dependent kinetic-based methods such as Trx-trapping mutants and thiol disulphide-independent methods such as BioID that we present here will bring us closer to completing this puzzle.
5. Conclusion
In this study we compared the applicability of two methods, BioID and immunoprecipitation with a crosslinker, for the identification of Prdx2 interactors in cells prior to lysis in a thiol-disulphide independent way. We found that tagging Prdx2 with a His-tag at the N-terminus significantly affects its peroxidase activity and possibly prevents its interaction with STAT3, which was not the case for the BioID construct. One of the interactors identified by BioID, subunit CSN5 from the COP9 signalosome, confirms earlier findings and could also be detected by PLA.
Hence, we demonstrate that BioID can be used as a method to find novel interactors of peroxiredoxins, and, potentially, other redox-active proteins. Our findings with His-tagged Prdx2 also suggest that caution should be exercised when interpreting the results obtained using tagged Prdxs in PPI studies.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
Ting Luo would like to express his gratitude for all the nights together with his beloved wonderful wife Jaitip Tipanee while editing figures. C.Y. thanks the European Space Agency (ESA) PRODEX program and the International Joint Research Group (IJRG) VUB-EPFL BioNanotechnology & NanoMedicine (NANO) for support in funding the Eclipse Ti2 Inverted Microscope (Nikon). We are grateful to Marc Fransen and Celien Lismont for critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102066.
Contributor Information
Daria Ezeriņa, Email: daria.ezerina@vub.be.
Joris Messens, Email: joris.messens@vub.be.
Funding
This work was funded with a VIB grant (to J.M.), and the Research Foundation-Flanders–Fonds de la Recherche Scientifique Excellence of Science project no. 30829584 (to J.M and D.V.). Ting L. was supported with a Chinese Scholarship Council grant (File No. 201707650018). C.Y. acknowledges FWO for the funding of her SB PhD fellowship (1S72218 N – 117745).
Contribution of the authors
Ting L., J.M. and D.E. designed the research. Ting L., J.M.P., K.W., C.Y., and D.V. performed experiments. Ting L., D.E., J.M., S.P.D.R and D.V. analysed the data. Tamas L. performed the bioinformatic analysis. Ting L., D.E., and J.M. wrote the paper. All authors read, edited, and agreed upon the final version of the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 2.Pedre B., Young D., Charlier D., Mourenza A., Rosado L.A., Marcos-Pascual L., Wahni K., Martens E., A G.d.l.R., Belousov V.V., Mateos L.M., Messens J. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11623–E11632. doi: 10.1073/pnas.1807954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barranco-Medina S., Lazaro J.J., Dietz K.J. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 4.Sun W., Dai L., Yu H., Puspita B., Zhao T., Li F., Tan J.L., Lim Y.T., Chen M.W., Sobota R.M., Tenen D.G., Prabhu N., Nordlund P. Monitoring structural modulation of redox-sensitive proteins in cells with MS-CETSA. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxidants Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skryhan K., Cuesta-Seijo J.A., Nielsen M.M., Marri L., Mellor S.B., Glaring M.A., Jensen P.E., Palcic M.M., Blennow A. The role of cysteine residues in redox regulation and protein stability of Arabidopsis thaliana starch synthase 1. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandes N., Schmitt S., Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxidants Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messina S., De Simone G., Ascenzi P. Cysteine-based regulation of redox-sensitive Ras small GTPases. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrer-Sueta G., Manta B., Botti H., Radi R., Trujillo M., Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 10.Winterbourn C.C. The biological chemistry of hydrogen peroxide. Methods Enzymol. 2013;528:3–25. doi: 10.1016/B978-0-12-405881-1.00001-X. [DOI] [PubMed] [Google Scholar]
- 11.Lim J.B., Huang B.K., Deen W.M., Sikes H.D. Analysis of the lifetime and spatial localization of hydrogen peroxide generated in the cytosol using a reduced kinetic model. Free Radic. Biol. Med. 2015;89:47–53. doi: 10.1016/j.freeradbiomed.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Stocker S., Van Laer K., Mijuskovic A., Dick T.P. The conundrum of hydrogen peroxide signaling and the emerging role of peroxiredoxins as redox relay hubs. Antioxidants Redox Signal. 2018;28:558–573. doi: 10.1089/ars.2017.7162. [DOI] [PubMed] [Google Scholar]
- 13.Fomenko D.E., Koc A., Agisheva N., Jacobsen M., Kaya A., Malinouski M., Rutherford J.C., Siu K.L., Jin D.Y., Winge D.R., Gladyshev V.N. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stocker S., Maurer M., Ruppert T., Dick T.P. A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol. 2018;14:148–155. doi: 10.1038/nchembio.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobotta M.C., Liou W., Stocker S., Talwar D., Oehler M., Ruppert T., Scharf A.N., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 16.Talwar D., Messens J., Dick T.P. A role for annexin A2 in scaffolding the peroxiredoxin 2-STAT3 redox relay complex. Nat. Commun. 2020;11:4512. doi: 10.1038/s41467-020-18324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis R.M., Hughes S.M., Ledgerwood E.C. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 18.van Dam L., Pages-Gallego M., Polderman P.E., van Es R.M., Burgering B.M.T., Vos H.R., Dansen T.B. The human 2-cys peroxiredoxins form widespread, cysteine-dependent- and isoform-specific protein-protein interactions. Antioxidants. 2021;10 doi: 10.3390/antiox10040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peskin A.V., Pace P.E., Behring J.B., Paton L.N., Soethoudt M., Bachschmid M.M., Winterbourn C.C. Glutathionylation of the active site cysteines of peroxiredoxin 2 and recycling by glutaredoxin. J. Biol. Chem. 2016;291:3053–3062. doi: 10.1074/jbc.M115.692798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 21.Beer S.M., Taylor E.R., Brown S.E., Dahm C.C., Costa N.J., Runswick M.J., Murphy M.P. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Santamarina S., Boronat S., Calvo I.A., Rodriguez-Gabriel M., Ayte J., Molina H., Hidalgo E. Is oxidized thioredoxin a major trigger for cysteine oxidation? Clues from a redox proteomics approach. Antioxidants Redox Signal. 2013;18:1549–1556. doi: 10.1089/ars.2012.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisz J.A., Bechtold E., King S.B., Poole L.B., Furdui C.M. Thiol-blocking electrophiles interfere with labeling and detection of protein sulfenic acids. FEBS J. 2013;280:6150–6161. doi: 10.1111/febs.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Netto L.E.S., Chae H.Z., Kang S.W., Rhee S.G., Stadtman E.R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J. Biol. Chem. 1996;271:15315–15321. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- 25.Sobotta M.C., Barata A.G., Schmidt U., Mueller S., Millonig G., Dick T.P. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic. Biol. Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Karala A.R., Ruddock L.W. Does s-methyl methanethiosulfonate trap the thiol-disulfide state of proteins? Antioxidants Redox Signal. 2007;9:527–531. doi: 10.1089/ars.2006.1473. [DOI] [PubMed] [Google Scholar]
- 27.Gotze M., Iacobucci C., Ihling C.H., Sinz A. A simple cross-linking/mass spectrometry workflow for studying system-wide protein interactions. Anal. Chem. 2019;91:10236–10244. doi: 10.1021/acs.analchem.9b02372. [DOI] [PubMed] [Google Scholar]
- 28.Wang H., He M., Willard B., Wu Q. Cross-linking, immunoprecipitation and proteomic analysis to identify interacting proteins in cultured cells. Bio Protoc. 2019;9 doi: 10.21769/BioProtoc.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner A., Joachimiak L.A., Unverdorben P., Walzthoeni T., Frydman J., Forster F., Aebersold R. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux K.J., Kim D.I., Burke B., May D.G. BioID: a screen for protein-protein interactions. Curr. Protein Pept. Sci. 2018;91:19 23 11–19 23 15. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V., Roux K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green N.M. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 33.Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mange A., Coyaud E., Desmetz C., Laurent E., Beganton B., Coopman P., Raught B., Solassol J. FKBP4 connects mTORC2 and PI3K to activate the PDK1/Akt-dependent cell proliferation signaling in breast cancer. Theranostics. 2019;9:7003–7015. doi: 10.7150/thno.35561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshinaka T., Kosako H., Yoshizumi T., Furukawa R., Hirano Y., Kuge O., Tamada T., Koshiba T. Structural basis of mitochondrial scaffolds by prohibitin complexes: insight into a role of the coiled-coil region. iScience. 2019;19:1065–1078. doi: 10.1016/j.isci.2019.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gheiratmand L., Coyaud E., Gupta G.D., Laurent E.M., Hasegan M., Prosser S.L., Goncalves J., Raught B., Pelletier L. Spatial and proteomic profiling reveals centrosome-independent features of centriolar satellites. EMBO J. 2019;38 doi: 10.15252/embj.2018101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zlatic S.A., Ryder P.V., Salazar G., Faundez V. Isolation of labile multi-protein complexes by in vivo controlled cellular cross-linking and immuno-magnetic affinity chromatography. JoVE. 2010;(37) doi: 10.3791/1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shenoy S.K., Drake M.T., Nelson C.D., Houtz D.A., Xiao K., Madabushi S., Reiter E., Premont R.T., Lichtarge O., Lefkowitz R. J. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J. Biol. Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 39.Liu G., Claret F.X., Zhou F., Pan Y. Jab1/COPS5 as a novel biomarker for diagnosis, prognosis, therapy prediction and therapeutic tools for human cancer. Front. Pharmacol. 2018;9:135. doi: 10.3389/fphar.2018.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longo P.A., Kavran J.M., Kim M.S., Leahy D.J. Transient mammalian cell transfection with polyethylenimine (PEI) Methods Enzymol. 2013;529:227–240. doi: 10.1016/B978-0-12-418687-3.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pak V.V., Ezerina D., Lyublinskaya O.G., Pedre B., Tyurin-Kuzmin P.A., Mishina N.M., Thauvin M., Young D., Wahni K., Martinez Gache S.A., Demidovich A.D., Ermakova Y.G., Maslova Y.D., Shokhina A.G., Eroglu E., Bilan D.S., Bogeski I., Michel T., Vriz S., Messens J., Belousov V.V. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metabol. 2020;31:642–653. doi: 10.1016/j.cmet.2020.02.003. e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hesketh G.G., Youn J.Y., Samavarchi-Tehrani P., Raught B., Gingras A.C. Parallel exploration of interaction space by BioID and affinity purification coupled to mass spectrometry. Methods Mol. Biol. 2017;1550:115–136. doi: 10.1007/978-1-4939-6747-6_10. [DOI] [PubMed] [Google Scholar]
- 43.Goemaere J., Knoops B. Peroxiredoxin distribution in the mouse brain with emphasis on neuronal populations affected in neurodegenerative disorders. J. Comp. Neurol. 2012;520:258–280. doi: 10.1002/cne.22689. [DOI] [PubMed] [Google Scholar]
- 44.Randall L.M., Dalla Rizza J., Parsonage D., Santos J., Mehl R.A., Lowther W.T., Poole L.B., Denicola A. Unraveling the effects of peroxiredoxin 2 nitration; role of C-terminal tyrosine 193. Free Radic. Biol. Med. 2019;141:492–501. doi: 10.1016/j.freeradbiomed.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ordonez E., Van Belle K., Roos G., De Galan S., Letek M., Gil J.A., Wyns L., Mateos L.M., Messens J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 2009;284:15107–15116. doi: 10.1074/jbc.M900877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedre B., Van Molle I., Villadangos A.F., Wahni K., Vertommen D., Turell L., Erdogan H., Mateos L.M., Messens J. The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Mol. Microbiol. 2015;96:1176–1191. doi: 10.1111/mmi.12998. [DOI] [PubMed] [Google Scholar]
- 47.Van Laer K., Buts L., Foloppe N., Vertommen D., Van Belle K., Wahni K., Roos G., Nilsson L., Mateos L.M., Rawat M., van Nuland N.A., Messens J. Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 2012;86:787–804. doi: 10.1111/mmi.12030. [DOI] [PubMed] [Google Scholar]
- 48.Schiapparelli L.M., McClatchy D.B., Liu H.H., Sharma P., Yates J.R., 3rd, Cline H.T. Direct detection of biotinylated proteins by mass spectrometry. J. Proteome Res. 2014;13:3966–3978. doi: 10.1021/pr5002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pace P.E., Peskin A.V., Konigstorfer A., Jasoni C.J., Winterbourn C.C., Hampton M.B. Peroxiredoxin interaction with the cytoskeletal-regulatory protein CRMP2: investigation of a putative redox relay. Free Radic. Biol. Med. 2018;129:383–393. doi: 10.1016/j.freeradbiomed.2018.10.407. [DOI] [PubMed] [Google Scholar]
- 50.Peskin A.V., Low F.M., Paton L.N., Maghzal G.J., Hampton M.B., Winterbourn C.C. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 51.Romanello K.S., Teixeira K.K.L., Silva J., Nagamatsu S.T., Bezerra M.A.C., Domingos I.F., Martins D.A.P., Araujo A.S., Lanaro C., Breyer C.A., Ferreira R.A., Franco-Penteado C., Costa F.F., Malavazi I., Netto L.E.S., de Oliveira M.A., Cunha A.F. Global analysis of erythroid cells redox status reveals the involvement of Prdx1 and Prdx2 in the severity of beta thalassemia. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Z., Bhella D., Lindsay J.G. Reconstitution of the mitochondrial PrxIII antioxidant defence pathway: general properties and factors affecting PrxIII activity and oligomeric state. J. Mol. Biol. 2007;372:1022–1033. doi: 10.1016/j.jmb.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Poole L.B., Nelson K.J. Distribution and features of the six classes of peroxiredoxins. Mol. Cell. 2016;39:53–59. doi: 10.14348/molcells.2016.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pastor-Flores D., Talwar D., Pedre B., Dick T.P. Real-time monitoring of peroxiredoxin oligomerization dynamics in living cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:16313–16323. doi: 10.1073/pnas.1915275117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Czupryna J., Tsourkas A Xanthine oxidase-generated hydrogen peroxide is a consequence, not a mediator of cell death. FEBS J. 2012;279:844–855. doi: 10.1111/j.1742-4658.2012.08475.x. [DOI] [PubMed] [Google Scholar]
- 57.Sakuma S., Abe M., Kohda T., Fujimoto Y. Hydrogen peroxide generated by xanthine/xanthine oxidase system represses the proliferation of colorectal cancer cell line Caco-2. J. Clin. Biochem. Nutr. 2015;56:15–19. doi: 10.3164/jcbn.14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gromer S., Arscott L.D., Williams C.H., Jr., Schirmer R.H., Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J. Biol. Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 59.Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 60.Bertolotti M., Bestetti S., Garcia-Manteiga J.M., Medrano-Fernandez I., Dal Mas A., Malosio M.L., Sitia R. Tyrosine kinase signal modulation: a matter of H2O2 membrane permeability? Antioxidants Redox Signal. 2013;19:1447–1451. doi: 10.1089/ars.2013.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johansson K., Cebula M., Rengby O., Dreij K., Carlstrom K.E., Sigmundsson K., Piehl F., Arner E.S. Cross talk in HEK293 cells between Nrf2, HIF, and NF-kappaB activities upon challenges with redox therapeutics characterized with single-cell resolution. Antioxidants Redox Signal. 2017;26:229–246. doi: 10.1089/ars.2015.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu L., Liu K., Ferreira R.B., Carroll K.S., Yang J. Proteome-wide analysis of cysteine S-sulfenylation using a benzothiazine-based probe. Curr. Protein Pept. Sci. 2019;95:e76. doi: 10.1002/cpps.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C., Kolas N., O'Donnell L., Leung G., McAdam R., Zhang F., Dolma S., Willems A., Coulombe-Huntington J., Chatr-Aryamontri A., Dolinski K., Tyers M. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Huo K., Ma L., Tang L., Li D., Huang X., Yuan Y., Li C., Wang W., Guan W., Chen H., Jin C., Wei J., Zhang W., Yang Y., Liu Q., Zhou Y., Zhang C., Wu Z., Xu W., Zhang Y., Liu T., Yu D., Zhang Y., Chen L., Zhu D., Zhong X., Kang L., Gan X., Yu X., Ma Q., Yan J., Zhou L., Liu Z., Zhu Y., Zhou T., He F., Yang X. Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deane C.M., Salwinski L., Xenarios I., Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 66.Guo Z., Wang Y., Zhao Y., Shu Y., Liu Z., Zhou H., Wang H., Zhang W. The pivotal oncogenic role of Jab1/CSN5 and its therapeutic implications in human cancer. Gene. 2019;687:219–227. doi: 10.1016/j.gene.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 67.Lambert J.P., Tucholska M., Go C., Knight J.D., Gingras A.C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bader J.S., Chaudhuri A., Rothberg J.M., Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 69.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 70.Rhee S.G., Woo H.A. Multiple functions of 2-Cys peroxiredoxins, I and II, and their regulations via post-translational modifications. Free Radic. Biol. Med. 2020;152:107–115. doi: 10.1016/j.freeradbiomed.2020.02.028. [DOI] [PubMed] [Google Scholar]
- 71.Xiao N., Du G., Frohman M.A. Peroxiredoxin II functions as a signal terminator for H2O2-activated phospholipase D1. FEBS J. 2005;272:3929–3937. doi: 10.1111/j.1742-4658.2005.04809.x. [DOI] [PubMed] [Google Scholar]
- 72.Chu K.L., Lew Q.J., Rajasegaran V., Kung J.T., Zheng L., Yang Q., Shaw R., Cheong N., Liou Y.C., Chao S.H. Regulation of PRDX1 peroxidase activity by Pin1. Cell Cycle. 2013;12:944–952. doi: 10.4161/cc.23916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parmigiani R.B., Xu W.S., Venta-Perez G., Erdjument-Bromage H., Yaneva M., Tempst P., Marks P.A. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Middaugh C.R., Vanin E.F., Ji T.H. Chemical crosslinking of cell membranes. Mol. Cell. Biochem. 1983;50:115–141. doi: 10.1007/BF00285638. [DOI] [PubMed] [Google Scholar]
- 75.Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May D.G., Scott K.L., Campos A.R., Roux K.J. Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells. 2020;9 doi: 10.3390/cells9051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belo Y., Mielko Z., Nudelman H., Afek A., Ben-David O., Shahar A., Zarivach R., Gordan R., Arbely E. Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine. Biochim. Biophys. Acta Gen. Subj. 2019;1863:1343–1350. doi: 10.1016/j.bbagen.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maity P.C., Yang J., Klaesener K., Reth M. The nanoscale organization of the B lymphocyte membrane. Biochim. Biophys. Acta. 2015;1853:830–840. doi: 10.1016/j.bbamcr.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antonicka H., Lin Z.Y., Janer A., Aaltonen M.J., Weraarpachai W., Gingras A.C., Shoubridge E.A. A high-density human mitochondrial proximity interaction network. Cell Metabol. 2020;32:479–497. doi: 10.1016/j.cmet.2020.07.017. e479. [DOI] [PubMed] [Google Scholar]
- 80.Go C.D., Knight J.D.R., Rajasekharan A., Rathod B., Hesketh G.G., Abe K.T., Youn J.Y., Samavarchi-Tehrani P., Zhang H., Zhu L.Y., Popiel E., Lambert J.P., Coyaud E., Cheung S.W.T., Rajendran D., Wong C.J., Antonicka H., Pelletier L., Palazzo A.F., Shoubridge E.A., Raught B., Gingras A.C. A proximity-dependent biotinylation map of a human cell. Nature. 2021;595:120–124. doi: 10.1038/s41586-021-03592-2. [DOI] [PubMed] [Google Scholar]
- 81.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mathys H., Basquin J., Ozgur S., Czarnocki-Cieciura M., Bonneau F., Aartse A., Dziembowski A., Nowotny M., Conti E., Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 83.Finci L.I., Zhang X., Huang X., Zhou Q., Tsai J., Teng T., Agrawal A., Chan B., Irwin S., Karr C., Cook A., Zhu P., Reynolds D., Smith P.G., Fekkes P., Buonamici S., Larsen N.A. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 2018;32:309–320. doi: 10.1101/gad.311043.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altmann E., Erbel P., Renatus M., Schaefer M., Schlierf A., Druet A., Kieffer L., Sorge M., Pfister K., Hassiepen U., Jones M., Ruedisser S., Ostermeier D., Martoglio B., Jefferson A.B., Quancard J. Azaindoles as zinc-binding small-molecule inhibitors of the JAMM protease CSN5. Angew Chem. Int. Ed. Engl. 2017;56:1294–1297. doi: 10.1002/anie.201608672. [DOI] [PubMed] [Google Scholar]
- 85.Ambroggio X.I., Rees D.C., Deshaies R.J. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hwang C.Y., Ryu Y.S., Chung M.S., Kim K.D., Park S.S., Chae S.K., Chae H.Z., Kwon K.S. Thioredoxin modulates activator protein 1 (AP-1) activity and p27Kip1 degradation through direct interaction with Jab1. Oncogene. 2004;23:8868–8875. doi: 10.1038/sj.onc.1208116. [DOI] [PubMed] [Google Scholar]
- 87.Gingras A.C., Abe K.T., Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 2019;48:44–54. doi: 10.1016/j.cbpa.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 88.Schopp I.M., Amaya Ramirez C.C., Debeljak J., Kreibich E., Skribbe M., Wild K., Bethune J. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017;8 doi: 10.1038/ncomms15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langford T.F., Huang B.K., Lim J.B., Moon S.J., Sikes H.D. Monitoring the action of redox-directed cancer therapeutics using a human peroxiredoxin-2-based probe. Nat. Commun. 2018;9:3145. doi: 10.1038/s41467-018-05557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chaudhuri R., Krycer J.R., Fazakerley D.J., Fisher-Wellman K.H., Su Z., Hoehn K.L., Yang J.Y.H., Kuncic Z., Vafaee F., James D.E. The transcriptional response to oxidative stress is part of, but not sufficient for, insulin resistance in adipocytes. Sci. Rep. 2018;8:1774. doi: 10.1038/s41598-018-20104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wei N., Serino G., Deng X.W. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Shackleford T.J., Zhang Q., Tian L., Vu T.T., Korapati A.L., Baumgartner A.M., Le X.F., Liao W.S., Claret F.X. Stat3 and CCAAT/enhancer binding protein beta (C/EBP-beta) regulate Jab1/CSN5 expression in mammary carcinoma cells. Breast Cancer Res. 2011;13:R65. doi: 10.1186/bcr2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bramasole L., Sinha A., Gurevich S., Radzinski M., Klein Y., Panat N., Gefen E., Rinaldi T., Jimenez-Morales D., Johnson J., Krogan N.J., Reis N., Reichmann D., Glickman M.H., Pick E. Proteasome lid bridges mitochondrial stress with Cdc53/Cullin1 NEDDylation status. Redox Biol. 2019;20:533–543. doi: 10.1016/j.redox.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou F., Pan Y., Wei Y., Zhang R., Bai G., Shen Q., Meng S., Le X.F., Andreeff M., Claret F.X. Jab1/Csn5-Thioredoxin signaling in relapsed acute monocytic leukemia under oxidative stress. Clin. Canc. Res. 2017;23:4450–4461. doi: 10.1158/1078-0432.CCR-16-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nicolussi A., D'Inzeo S., Capalbo C., Giannini G., Coppa A. The role of peroxiredoxins in cancer. Mol Clin Oncol. 2017;6:139–153. doi: 10.3892/mco.2017.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.