Abstract
Objectives
To analyse the context of genes conferring antibiotic resistance in two carbapenem-resistant Acinetobacter baumannii isolates recovered in Tehran, Iran.
Methods
The antibiotic resistance phenotype for 28 antibiotics was determined using disc diffusion. The whole genome sequences of ABH008 and ABS200 were determined using the Illumina HiSeq X Ten platform. Resistance genes were identified using ResFinder and multilocus sequence types were determined using the Oxford and Institut Pasteur schemes.
Results
Isolates ABH008 and ABS200, recovered in 2012 and 2013, respectively, in two different Tehran hospitals, belong to the common global clone 1 lineage, ST1IP and ST231OX. They are resistant to sulfamethoxazole, tetracycline, gentamicin, amikacin, third-generation cephalosporins and carbapenems. Despite being isolated in different hospitals, phylogenetic analysis indicated they are closely related. Consistent with this, both isolates carry catA1, sul1, aacC1 and aadA1 in a novel variant of the AbaR3-type resistance island, named AbaR31. Both isolates are resistant to amikacin and carbapenems owing to aphA6 and oxa23, respectively. The oxa23 gene is located in the AbaR4 resistance island, and aphA6 in TnaphA6, and both mobile elements are in an ∼90 kbp plasmid encoding the putative RepAci6 replication initiation protein. Resistance to third-generation cephalosporins is due to the acquisition by homologous recombination of a 5 kb DNA segment that contains ISAba1-ampC from a ST623 strain.
Conclusions
The resistance gene complements of ABH008 and ABS200 were found in AbaR31 and a plasmid that encodes RepAci6. The close genetic relationship of ABH008 and ABS200, despite each being recovered from different hospitals, indicates transmission between the two hospitals.
Introduction
Resistance to carbapenems in Acinetobacter baumannii, the number one WHO priority pathogen, is widespread, posing a global threat as carbapenems are the frontline option for treating infections caused by multi-antibiotic-resistant strains of this opportunistic pathogen.1,2 In A. baumannii carbapenem resistance is most often caused by acquired class D carbapenem oxacillinases, typically OXA-23, OXA-24 (OXA-40) and OXA-58.2,3
Two clonal complexes of A. baumannii are distributed globally: global clone 1 (GC1) and global clone 2 (GC2). Within each is a myriad of lineages and sub-lineages. The genetic variation among these lineages informs understanding of how resistance evolves and enables epidemiological tracking of antibiotic-resistant strains. For A. baumannii, the majority (70%) of the whole genome sequence data feeding variant analysis is geographically skewed.2 It is sourced from isolates obtained from only four countries, all outside the Middle East region2 while the source of many resistant lineages of A. baumannii, however, is believed to be the Middle East region.4–7 This discrepancy makes it difficult to draw an accurate global picture of A. baumannii. We recently reported analysis of whole genome sequence data from an outbreak in an Iranian hospital that supports the view that lineage 2 GC1 strains may have an origin in the region.5
Here, we report analysis of whole genome sequence data for two antibiotic-resistant A. baumannii strains isolated from two different hospitals in Iran. Both belong to lineage 1 of GC1 indicating that strains belonging to lineage 1 are also present in the country.
Methods
Antibiotic resistance phenotype and PCR mapping
Antibiotic resistance profiles of ABS200 and ABH008 were determined using the standard Kirby-Bauer disc diffusion assay to 28 antibiotics as previously described.5 Resistance to colistin was determined by measuring the MICs using the standard micro-broth dilution method.8 Antibiotic discs tested were ampicillin, streptomycin, spectinomycin, sulfamethoxazole, tetracycline, trimethoprim, kanamycin, neomycin, cefotaxime, ceftazidime, gentamicin, ciprofloxacin, amikacin, nalidixic acid, tobramycin, netilmicin, imipenem, meropenem, ticarcillin/clavulanic acid, rifampicin, ampicillin/sulbactam, cefepime, doripenem, piperacillin/tazobactam, ceftriaxone, minocycline, doxycycline and levofloxacin. Strains were classified as resistant or susceptible according to the CLSI guidelines for Acinetobacter spp.9 and Calibrated Dichotomous Sensitivity (CDS) disc diffusion assay (http://web.med.unsw.edu.au/cdstest) where an Acinetobacter spp. CLSI breakpoint was not available (e.g. for netilmicin, streptomycin, spectinomycin, sulfamethoxazole, nalidixic acid and rifampicin).
AbaR resistance islands were mapped using the PCR mapping strategy previously developed including the novel AbaR31 junction, which was mapped using the RH513: 5′-GTACTGTTGTAATTCATTAAGCAT-3′ and intI1-RV: 5′-GGGCATGGTGGCTGAAGGACC-3′ primers generating a 3 kb product.10,11
AbaR4 was also mapped using previously designed primers12 and located in a RepAci6 plasmid using two mapping PCRs (i) MH5 (5′-CTAAAAGGCGTTTGGGCATA-3′) and RH909 (5′-GCGATTCAAAATATCGGTCAA-3′)13 (generating a 3105 bp product) and (ii) MH7 (5′-TGCTGAACCGTACAACCAGA-3′) and MH6 (5′-CAGGATGAGCTGGATCAACA-3′) (generating a 3645 bp product) developed here. PCR conditions were the same as those described elsewhere.
Whole genome sequencing, assembly and phylogenetic analysis
Genomic DNA isolated from ABH008 and ABS200 was sequenced using Illumina HiSeq X Ten. Paired-end reads of 150 bp were assembled using SPAdes (v. 3.14.1) yielding 75 and 77 contigs for ABH008 and ABS200, respectively, and with an average depth of ∼100-fold for each genome. Multi-locus Sequence Types in the Institut Pasteur (STIP) and Oxford (STOX) schemes as well as the ampC alleles were determined from the genome sequence data using the PubMLST database (http://pubmlst.org/abaumannii/).
Antibiotic resistance genes and contigs carrying them were identified using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/), and recovered using standalone BLAST (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/).
Genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (v.4.11).14 Pfam (http://pfam.xfam.org/) and ISFinder (https://www-is.biotoul.fr/) search tools were used to identify proteins and insertion sequences, respectively. Here, we used the STIP#:STOX#:KL#:OCL# formula that we recently developed15 to group/distinguish A. baumannii isolates.
Phylogenetic analysis was done by constructing a recombination-free tree by generating a whole genome alignment using snippy (v.4.6.0) (publicly available at https://github.com/tseemann/snippy) followed by removing regions potentially introduced by recombination using Gubbins16 (v2.1.025) as previously described in detail.5,6 A set of nine GC1 strains known to belong to lineage 1 and 2, including the A1 GC1 reference genome (GenBank accession numberCP010781), were also used as controls (Figure 1). Figures were drawn to scale using SnapGene® (v. 5.2.4.) and Inkskape (v. 1.0).
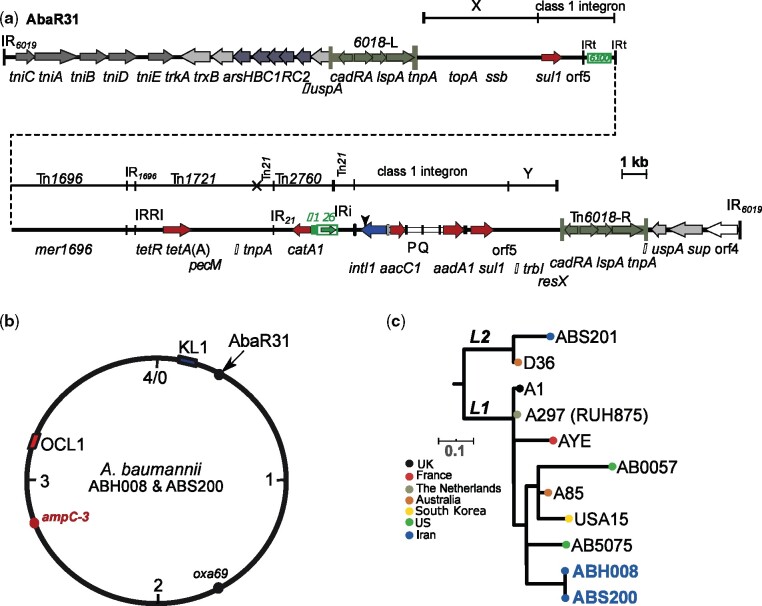
Figure 1.
Structure of AbaR31 (a), circular map representing the chromosomes of ABH008 and ABS200 (b) and phylogenetic tree of representative GC1 strains (c). In (a), The thick central line shows the AbaR31 resistance island with horizontal arrows indicating the extent and orientation of genes. Antibiotic resistance genes are shown using red arrows. Two terminal vertical lines indicate the inverted repeats (IRs) of AbaR31. Tn6018 copies are shown using dark green. The origin of each segment of the multiple antibiotic resistance region (MARR), flanked by Tn6018 copies, is shown using a thin line above the central line. The vertical arrowhead above the intI1 gene indicates the 108 bp deletion found in AbaR31. (b) Represents a schematic of the ABH008 and ABS200 chromosome with the position of AbaR31, ampC, K and OC loci marked. (c) Shows the phylogenetic relationship of ABH008 and ABS200 compared with known GC1 strains including GC1 reference strain A297 (RUH875).
Nucleotide accession numbers
The genome sequence of ABH008 and ABS200 have been deposited in GenBank GenBank/EMBL/DDBJ database and are publicly available under the accession numbers JADPVA000000000 and JADPVB000000000.
Results and discussion
Resistance profiles, MLST and phylogenetic relationship
ABH008 and ABS200 are carbapenem-resistant isolates from two different hospitals in Tehran, Iran, isolated in 2012 and 2013, respectively. We determined their resistance phenotype and their relationship to global A. baumannii clones. ABH008 and ABS200 are resistant to carbapenems, third-generation cephalosporins, ticarcillin/clavulanic acid, tetracycline, streptomycin, spectinomycin, gentamicin, kanamycin, neomycin, amikacin sulphonamides, trimethoprim, nalidixic acid and ciprofloxacin, (Table S1, available as Supplementary data at JAC-AMR Online) while remaining susceptible to colistin (MIC <0.5 mg/L). Hence, they are considered extensively drug resistant (XDR).17 ABH008 and ABS200 genomes both contain the catA1, tetA(A), sul1, aacC1, aadA1, aphA6 and oxa23 genes, accounting for the resistance phenotype observed. ABH008 and ABS200 were found to be GC1 based on analysis of the allelic-specific PCR18 which determines clones and their draft genomes. Consistent with other GC1s, both strains encode the OXA-69 variant of the intrinsic oxaAb gene and belong to ST1IP(1-1-1-1-1-5-1) and ST231OX(10-12-4-11-4-98-5). Phylogenetic analysis showed ABH008 and ABS200 were tightly clustered together, both falling within lineage 1 (Figure 1). It is possible that the ABH008 and ABS200 branch might represent the lineage 1 strains circulating in Tehran’s hospitals; however, more genome sequences would be needed to confirm this.
ABH008 and ABS200 carry AbaR31; a novel antibiotic resistance island derived from AbaR3
To determine the genetic context of the resistance genes of ABH008 and ABS200, we used a combination of draft genomes and PCR mapping strategies.10–13 The catA1, tetA(A), sul1, aacC1, aadA1 resistance genes were localized in a novel variant of AbaR0/3-type islands with a Tn6019 transposon backbone located in the comM gene of ABH008 and ABS200. Our draft genome analysis indicated that this island derives from AbaR3 rather than AbaR0 as it contains a 108 bp sequence in the 5′-CS of the class 1 integron, which is characteristic of AbaR3 (e.g. in AB0057; GenBank accession numberCP001182) and its variants.19 We named this new AbaR3 variant AbaR31. It is predicted to be 51 113 kbp (Figure 1). AbaR31 has been generated via an IS26-mediated adjacent deletion of an 11 877 bp internal segment of the AbaR3 island that includes the blaTEM and aphA1b antibiotic resistance genes. IS26-mediated deletions are known to be important in generating variants of AbaR0/3 islands.19 We confirmed the novel junction in AbaR31 generated by the IS26-mediated deletion by manual sequencing of the ∼3 kbp product of a PCR that links intI1 with catA1. The presence of AbaR31 in both ABH008 and ABS200 indicates their close relationship as well as transmission between hospitals.
AbaR4 and TnaphA6 carried on a RepAci6 plasmid
The two antibiotic resistance genes of ABH008 and ABS200, aphA6 and oxa23, that are not carried on the AbaR31 island were found in different transposons on the same plasmid. Using published primers13 we previously showed that the aphA6 amikacin resistance gene is located in TnaphA6 inABH008 and ABS200 in a context similar to that seen in RepAci6 plasmids.20 Here, analysis of draft genomes confirmed that the aphA6 gene is in TnaphA6, in precisely the location that this transposon is in pAb-G7-2 (GenBank accession numberKF669606).13 The plasmid pAb-G7-2 is a RepAci6 plasmid found in an Australian GC1.13 All remaining segments of pAb-G7-2 were recovered in multiple contigs, indicating that the RepAci6 plasmid in ABH008 and ABS200 is closely related to pAb-G7-2 (with 99.9% DNA identities in plasmid backbone). The oxa23 gene is in Tn2006 in AbaR4; this was confirmed by retrieving all of the contigs making up the complete AbaR4 from draft genomes. Using the genome sequence data, AbaR4 was predicted to be located on the same RepAci6 plasmid that contains TnaphA6. This prediction was confirmed by developing a PCR mapping strategy, followed by manual sequencing of products, to link unique sequences of either end of AbaR4 (tniB on the left and oxa23 on the right) to the backbone of RepAci6 plasmid (see Methods). This led to the localization of AbaR4 in a spot equivalent to the position 9601 of pAb-G7-2 (GenBank accession numberKF669606) interrupting an open reading frame that encodes a hypothetical protein (protein id AGY56207.1). Sequence analysis also confirmed that AbaR4 is flanked by 5 bp target site duplication (TSD) 5′-AAAAG-3′, a typical property of transposons that belong to this family.12 AbaR4 has already been detected in RepAci6 plasmids, e.g. pA85-3 (GenBank accession numberCP021787); however, in the genomes of ABH008 and ABS200 it is present in a unique location in the plasmid backbone, which indicates local acquisition is likely. RepAci6 plasmids are potentially conjugative,13,21–23 and have been associated with the aphA6 (amikacin resistance) and oxa23 (carbapenem resistance) genes singly,6,24 or together,23 which combined makes RepAci6 plasmids important vehicles to spread amikacin and carbapenem resistance genes. To date, RepAci6 plasmids have not been seen in other Gram-negative bacteria e.g. enterobacteria or Klebsiella pneumoniae.
Analysis of the draft genomes of ABH008 and ABS200 also showed both strains contain a copy of a cryptic plasmid encoding RepAci1 that was identical to pA1-1 (GenBank accession numberCP010782) found in A. baumannii A1 recovered in the UK in 1982.25 The plasmid pA1-1 and its variants encode the AbkA (pfam14384) and AbkB (pfam04365) toxin–antitoxin system, which helps plasmids to be stably maintained in the cells. This plasmid type is widespread in A. baumannii strains regardless of clonal type and geographical distribution.6,24
Resistance to third-generation cephalosporins due to acquisition of a DNA segment containing ISAba1-ampC
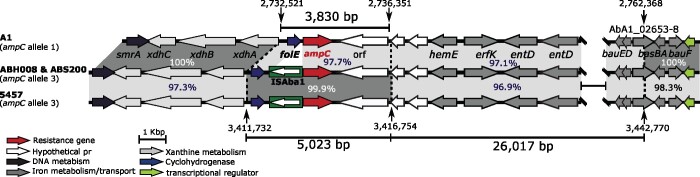
The presence of ISAba1 upstream of the intrinsic ampC gene is known to be responsible for increasing the expression level of this gene, leading to resistance to third-generation cephalosporins.26 Both ABH008 and ABS200 include a copy of ISAba1 9 bp away from the start of ampC, explaining their resistance to ceftazidime and cefotaxime. However, analysis of the ampC sequences of ABH008 and ABS200 revealed that they do not include the standard ST1 ampC sequence (allele 1). Instead, they carry the allele 3 ampC sequence, typical in ST623 strains (e.g. strain 5457; GenBank accession numberCP045541). We have previously reported several cases of the acquisition of DNA segments containing an ISAba1-activated ampC gene by homologous recombination leading to third-generation cephalosporin resistance.27,28 Similarly, comparative analysis of the ampC region in ABH008 and ABS200 with the corresponding regions in A1 (GenBank accession numberCP010781 containing allele 1) and 5457 (GenBank accession numberCP045541 containing allele 3) also indicated that the ISAba1-ampC structure has been acquired from 5457 and incorporated into their chromosome as part of an ∼5 kbp DNA segment (Figure 2). A ∼26 kbp DNA patch was also found immediately adjacent to the 5 kbp fragment that contains ISAba1-ampC (Figure 2). This segment differed from the corresponding regions in A1 and 5457 by 97.1% and 96.9%, respectively (Figure 2). The source for this 26 kbp chromosomal segment could not be found. However, interestingly, an identical 5022 bp ampC-3 patch, along with its adjacent ∼26 kbp segment, was also found in the A. baumannii strain DA33382 (GenBank accession numberCP030106), indicating it is closely related to ABH008 and ABS200. DA33382 is an ST1:ST1567:KL40:OCL2 GC1 strain recovered from a tracheal secretion sample in Germany with an unknown isolation date. The fact that DA33382 has a different KL and OCL, compared with ABH008 and ABS200, indicates that it has diverged by switching its outer core and capsular surface polysaccharides. We previously proposed that the Middle East region might be the source for many of the globally distributed resistance lineages of GC1s.5,6 The relationship of DA33382, ABH008 and ABS200 provides further evidence of this, but more genomes from the region will be needed to confirm the proposition.
Figure 2.
Schematic representation of the ampC region in ABH008 and ABS200 compared with A1 (GenBank accession numberCP010781) and 5457 (GenBank accession numberCP045541). The thick central line shows the chromosomes and horizontal arrows indicate the extent and orientation of genes. Shades of grey indicate regions with significant identity and numbers show their percentage DNA identity. Vertical arrows with numbers (above or below them) indicate chromosomal positions of A1 and 5457 as in CP010781 and CP045541, respectively.
Conclusions
The ST1:ST231:KL1:OCL1 strains ABH008 and ABS200 belong to lineage 1 within global clone 1. Both strains are extensively antibiotic resistant due to the presence of AbaR31 and a RepAci6 plasmid that carries AbaR4 and TnaphA6. ABH008 and ABS200 show that strains belonging to lineage 1 are circulating in Tehran hospitals and that they are closely related to a strain from Germany.
Supplementary Material
Acknowledgements
We would like to thank Dr Fiona Mclver, of the University of Technology Sydney, Australia, for critical reading of this manuscript.
Funding
M.H. and the bioinformatics parts of this work were supported by an Australian Research Council (ARC) DECRA fellowship (fellowship DE200100111). M.D. and the experimental part of this study were supported by a Tehran University of Medical Sciences Grant (no. 41448) and the Academy of Science for Developing World (TWAS) Grant (no. 11-119 RG/PHA/AS_C-UNESCO FR:3240262646).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC-AMR Online.
References
- 1. WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017: 1–7. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
- 2. Hamidian M, Nigro SJ.. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 2019; 5: e000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poirel L, Naas T, Nordmann P.. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 2010; 54: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams MD, Goglin K, Molyneaux N. et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 2008; 190: 8053–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douraghi M, Kenyon JJ, Aris P. et al. Accumulation of antibiotic resistance genes in carbapenem-resistant Acinetobacter baumannii isolates belonging to lineage 2. Global Clone 1, from Outbreaks in 2012-2013 at a Tehran Burns Hospital. mSphere 2020; 5: e00164–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holt K, Kenyon JJ, Hamidian M. et al. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom 2016; 2: e000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hujer KM, Hujer AM, Hulten EA. et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 2006; 50: 4114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2001; 48: 5–16. [DOI] [PubMed] [Google Scholar]
- 9. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Ninth Edition: M100. 2019.
- 10. Post V, Hall RM.. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother 2009; 53: 2667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Post V, White PA, Hall RM.. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 2010; 65: 1162–70. [DOI] [PubMed] [Google Scholar]
- 12. Hamidian M, Hall RM.. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 2011; 66: 2484–91. [DOI] [PubMed] [Google Scholar]
- 13. Hamidian M, Holt KE, Pickard D. et al. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 2014; 69: 955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tatusova T, DiCuccio M, Badretdin A. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 2016; 44: 6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamidian M, Ambrose SJ, Blackwell GA. et al. An outbreak of multiply antibiotic-resistant ST49:ST128:KL11:OCL8 Acinetobacter baumannii isolates at a Sydney hospital. J Antimicrob Chemother 2021; 76: 893–900. [DOI] [PubMed] [Google Scholar]
- 16. Croucher NJ, Page AJ, Connor TR. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 18. Turton JF, Gabriel SN, Valderrey C. et al. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 2007; 13: 807–15. [DOI] [PubMed] [Google Scholar]
- 19. Hamidian M, Hall RM.. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 – structure, origin and evolution. Drug Resist Updat 2018; 41: 26–39. [DOI] [PubMed] [Google Scholar]
- 20. Aris P, Boroumand MA, Douraghi M.. Amikacin resistance due to the aphA6 gene in multi-antibiotic resistant Acinetobacter baumannii isolates belonging to global clone 1 from Iran. BMC Microbiol 2019; 19: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamidian M, Hall RM.. pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J Antimicrob Chemother 2014; 69: 1146–8. [DOI] [PubMed] [Google Scholar]
- 22. Hamidian M, Kenyon JJ, Holt KE. et al. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother 2014; 69: 2625–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nigro SJ, Holt KE, Pickard D. et al. Carbapenem and amikacin resistance on a large conjugative Acinetobacter baumannii plasmid. J Antimicrob Chemother 2015; 70: 1259–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hamidian M, Hawkey J, Wick R. et al. Evolution of a clade of Acinetobacter baumannii global clone 1, lineage 1 via acquisition of carbapenem- and aminoglycoside-resistance genes and dispersion of ISAba1. Microb Genom 2019; 5: e000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holt KE, Hamidian M, Kenyon JJ. et al. Genome sequence of Acinetobacter baumannii strain A1, an early example of antibiotic-resistant global clone 1. Genome Announc 2015; 3: e00032-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hamidian M, Hall RM.. ISAba1 targets a specific position upstream of the intrinsic ampC gene of Acinetobacter baumannii leading to cephalosporin resistance. J Antimicrob Chemother 2013; 68: 2682–3. [DOI] [PubMed] [Google Scholar]
- 27. Hamidian M, Hall RM.. Resistance to third-generation cephalosporins in Acinetobacter baumannii due to horizontal transfer of a chromosomal segment containing ISAba1-ampC. J Antimicrob Chemother 2014; 69: 2865–6. [DOI] [PubMed] [Google Scholar]
- 28. Hamidian M, Hancock DP, Hall RM.. Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J Antimicrob Chemother 2013; 68: 244–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.