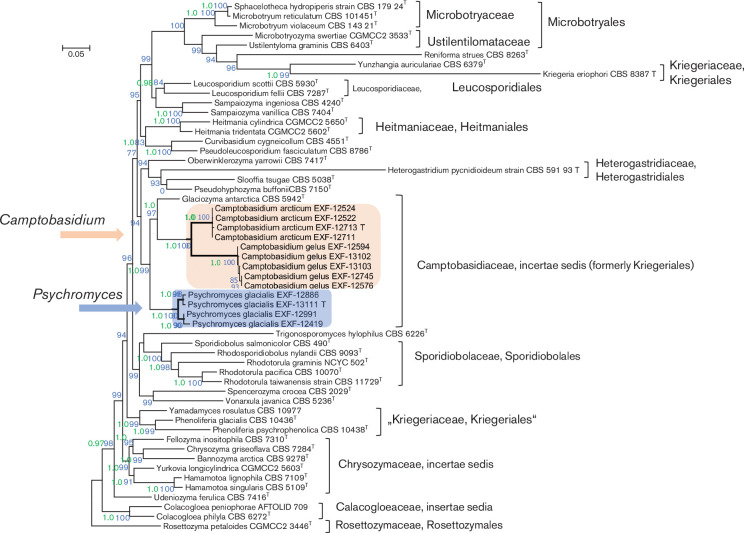
Fig. 2.
Phylogenetic tree based on alignment of the SSU, LSU, 5.8S rDNA, TEF, CYTB, RPB1 and RPB2 estimated by maximum-likelihood and Bayesian analyses. The tree includes representatives of Psychromyces glacialis gen. nov., sp. nov., Camptobasidium arcticum sp. nov., C. gelus and representative species of genera of Microbotryomycetes. In the maximum-likelihood estimation, the best model of nucleotide substitution was estimated with jModelTest, other parameters (alpha parameter of the gamma distribution of substitution rate categories, proportion of invariable sites) were estimated within PhyML. aLRT as Chi2-based support was used for calculation of branch supports. In estimation by MrBayes, a total 10 million generations were calculated and the first 25 % were discarded before the shown consensus tree was calculated from trees sampled every 100 generations. Posterior probabilities higher than 0.9 (in green) and maximum-likelihood bootstrap value from 1000 bootstrap replicates larger than 70 % (in blue) are shown near the nodes.