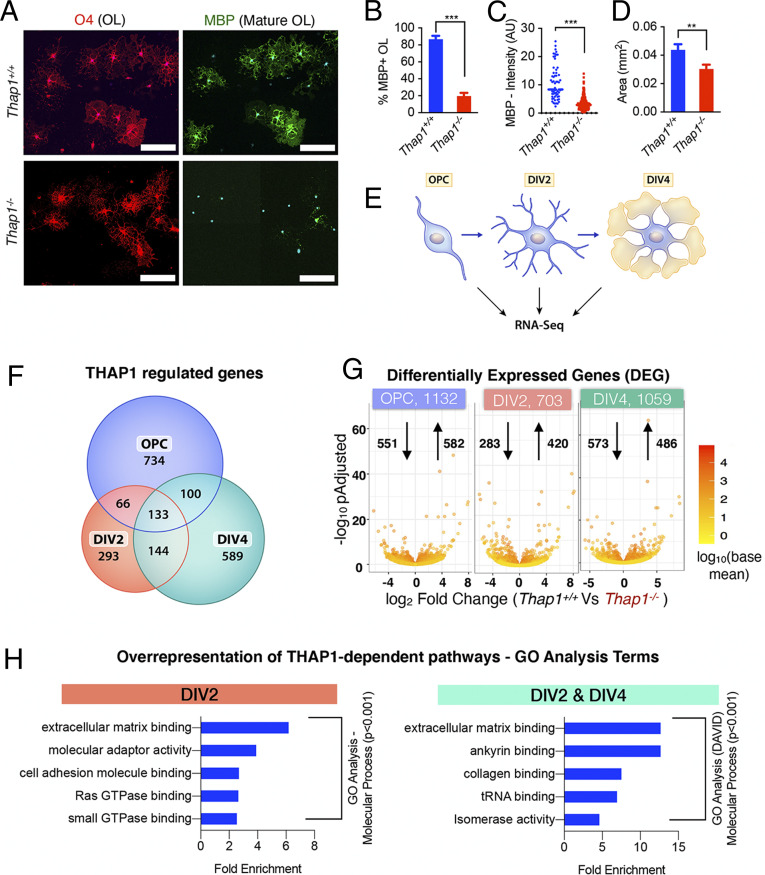
Fig. 1.
THAP1 loss disrupts the ECM transcriptome. (A–D) Loss of THAP1 causes maturation deficits in differentiating OPC. (A) Representative images of control (Thap1+/+) and THAP1 null (derived from Thap1flx/−;Olig2-Cre+) OL differentiated (+T3) for 4 d (scale bar, 100 μm.) Cultures are stained for O4, MBP, and DAPI. (B) Quantification of the percentage of O4+ cells expressing MBP. The bar graph shows mean values ± SEM Thap1+/+ = 86.78% ± 3.98; Thap1−/− = 19.58% ± 3.88; t test: t(4) = 12.07; P = 0.0003; and n = 3 clonal lines (100 cells per clone) per genotype. (C) Mean intensity of MBP staining in O4+ cells, represented as arbitrary units (AUs), normalized to background. Each data point in the scatter plot represents MBP+ staining intensity for individual cells. Thap1+/+ = 10.76 AU ± 0.76; Thap1−/− = 3.32 AU ± 0.108; t test: t(427) = 18.21; P < 0.0001; and n = 3 clonal lines (>100 cells per clone) per genotype. (D) Average area of O4+ cells. The bar graph shows mean values ± SEM Thap1+/+ = 0.043 mm2 ± 0.00402; Thap1−/− = 0.030 mm2 ± 0.00309; t test: t(633) = 2.639; P = 0.008; and n = 3 clonal lines (>100 cells per clone) per genotype. (E) Schematic illustrating RNAseq analyses. Total RNA was extracted from three independent clonal lines, each of Thap1+/+ and Thap1−/− cells, at three different time points (OPC, DIV2, and DIV4) and analyzed using Illumina Hiseq (Materials and Methods). (F) Venn diagram depicting the overlap of Thap1+/+ versus Thap1−/− DEGs from OPC, DIV2, and DIV4. (G) Number of DEGs at each time point analyzed. The x-axis = log2 fold change (magnitude of change for the DEG) in Thap1−/−. The y-axis = −log10 adjusted P value depicts the statistical significance of the change. The number of up- and down-regulated genes are indicated by arrows. (H) The DEG at either DIV2 (703) or both DIV2 and DIV4 (277) were used for enrichment analysis of GO terms to identify overrepresented biological pathways using both GENEONTOLOGY and DAVID. Graphs show the most significantly overrepresented GO terms (P < 0.001, sorted as fold overrepresented) in OL from DIV2 (GENEONTOLOGY, GO–Molecular Function) and from the DIV2 and DIV4 overlapping group (DAVID, GO–Molecular Function). tRNA, transfer RNA.