Significance
Novel antibiotics are urgently needed to resolve the current antimicrobial resistance crisis. For critical pathogens, drug entry through the cell envelope is one of the major challenges in the development of effective novel antibiotics. Envelope proteins forming water-filled channels, so-called porins, are commonly thought to be essential for entry of hydrophilic molecules, but we show here for the critical pathogen Pseudomonas aeruginosa that almost all antibiotics and diverse hydrophilic nutrients bypass porins and instead permeate directly through the outer membrane lipid bilayer. However, carboxylate groups hinder bilayer penetration, and Pseudomonas thus needs porins for efficient utilization of carboxylate-containing nutrients such as succinate. The major porin-independent entry route might open opportunities for facilitating drug delivery into bacteria.
Keywords: membrane transport, bacterial outer membrane, lipid bilayer, diffusion, antimicrobial resistance
Abstract
Gram-negative bacterial pathogens have an outer membrane that restricts entry of molecules into the cell. Water-filled protein channels in the outer membrane, so-called porins, facilitate nutrient uptake and are thought to enable antibiotic entry. Here, we determined the role of porins in a major pathogen, Pseudomonas aeruginosa, by constructing a strain lacking all 40 identifiable porins and 15 strains carrying only a single unique type of porin and characterizing these strains with NMR metabolomics and antimicrobial susceptibility assays. In contrast to common assumptions, all porins were dispensable for Pseudomonas growth in rich medium and consumption of diverse hydrophilic nutrients. However, preferred nutrients with two or more carboxylate groups such as succinate and citrate permeated poorly in the absence of porins. Porins provided efficient translocation pathways for these nutrients with broad and overlapping substrate selectivity while efficiently excluding all tested antibiotics except carbapenems, which partially entered through OprD. Porin-independent permeation of antibiotics through the outer-membrane lipid bilayer was hampered by carboxylate groups, consistent with our nutrient data. Together, these results challenge common assumptions about the role of porins by demonstrating porin-independent permeation of the outer-membrane lipid bilayer as a major pathway for nutrient and drug entry into the bacterial cell.
Antimicrobial resistance is a major worldwide threat to human health. The World Health Organization has classified Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii as the most concerning pathogens (“critical priority”) (1). All three pathogens are Gram-negative bacteria with the characteristic inner and outer membranes. The outer membrane is a stringent permeability barrier that restricts the entry of most molecules and therefore presents a major challenge for the development of urgently needed novel antibiotics (2–5).
The outer membrane consists of an asymmetric lipid bilayer with lipopolysaccharide (LPS) in the outer leaflet and phospholipids in the inner leaflet and various outer-membrane proteins that are embedded in, or attached to, the lipid bilayer. LPS contains negatively charged phosphate and carboxylate groups that are cross-linked by divalent Mg2+ and Ca2+ cations, resulting in stable clusters of LPS molecules that reduce the permeation of small molecules by 10- to 100-fold compared to phospholipid bilayers (6). Some outer membrane proteins form water-filled channels (so-called porins) that facilitate translocation of molecules through the outer membrane (4, 5). Enterobacteriaceae have general “unspecific” porins that permit the entry of molecules with a size of up to 600 Da. By contrast, P. aeruginosa and A. baumannii have a large set of “specific” porins that permit the entry of only few molecules with sizes below 200 Da. In addition, all three pathogens have porins with mainly structural roles in stabilizing the link between outer membrane and the underlying peptidoglycan layer (OmpA and OprF). It has been proposed that a small fraction of these structural porin molecules form large unspecific pores that permit entry of larger molecules at low rates (7), but this model remains controversial.
Antimicrobials and nutrients can penetrate the outer membrane by two different pathways, through the lipid bilayer or through porins. Hydrophobic molecules might predominantly use the lipid pathway, while hydrophilic molecules might prefer porins. However, the quantitative relevance of each pathway for outer-membrane permeability remains unknown (3, 8, 9). Even slow permeation pathways that mediate concentration-equilibration times in the order of minutes (instead of seconds) can yield relevant intracellular drug concentrations in bacteria with generation times of more than 20 min, unless drug-efflux pumps and/or hydrolases diminish drug levels (2).
Translocation pathways and their selectivity for specific physicochemical properties of molecules are crucial for the rational improvement of drug entry into Gram-negative bacteria. The important contribution of large cation-selective porins such as OmpF and OmpC for outer-membrane translocation into Enterobacteriaceae enabled the establishment of rules for medicinal chemistry to improve whole-cell activities of antimicrobials against these bacteria (10–12). These porins have been extensively studied, and in particular OmpF has a major impact on susceptibility to various β-lactam antibiotics (13). However, an Escherichia coli ΔompC ΔompF double mutant retains substantial susceptibility to diverse other antibiotics (9), suggesting alternative translocation pathways.
For P. aeruginosa, physicochemical parameters favoring translocation have been more difficult to identify (10, 14, 15). Both P. aeruginosa and A. baumannii have lower outer-membrane permeability than Enterobacteriaceae for hydrophilic molecules because they lack unspecific porins (16), making antimicrobial development particularly difficult for these critical pathogens. Specific porins might facilitate antibiotic entry into P. aeruginosa (17), but clear evidence for standard assay conditions is only available for penetration of carbapenems through OprD (18). Functional studies of individual porins in P. aeruginosa are hampered by the large diversity of specific porins that are thought to each enable uptake of a few nutrients (19). Phenotypes of inactivating one particular porin might be masked by the numerous remaining other porins. To circumvent these issues, individual porins have been purified and reconstituted in artificial membranes, or expressed in E. coli, to determine their substrate specificity. However, the results might not reflect porin functions in their native context because their channel properties differ depending on the lipid environment (20, 21).
In this study, we overcame these difficulties using extensive mutagenesis. In contrast to previous assumptions, we show that wild-type P. aeruginosa PA14 and a PA14 Δ40 mutant that lacks all identifiable 40 porin genes have indistinguishable susceptibility to diverse antibiotics. Moreover, the Δ40 strain grew normally on rich media, and nutrient consumption assays revealed substantial porin-independent uptake of diverse hydrophilic nutrients. Bringing back individual porins accelerated uptake of some neutral/zwitterionic molecules and was essential for efficient consumption of negatively charged carboxylate-containing compounds. Instead of narrow substrate specificity, porins actually had broad overlapping substrate selectivity. These results demonstrate an unexpected but efficient porin-independent translocation pathway through the outer-membrane lipid bilayer for diverse hydrophilic compounds and all antipseudomonal antibiotics. A detailed understanding of this pathway will facilitate the development of novel antibiotics.
Results
The “General” Porin OprF Has Limited Relevance for Translocation of Antimicrobials.
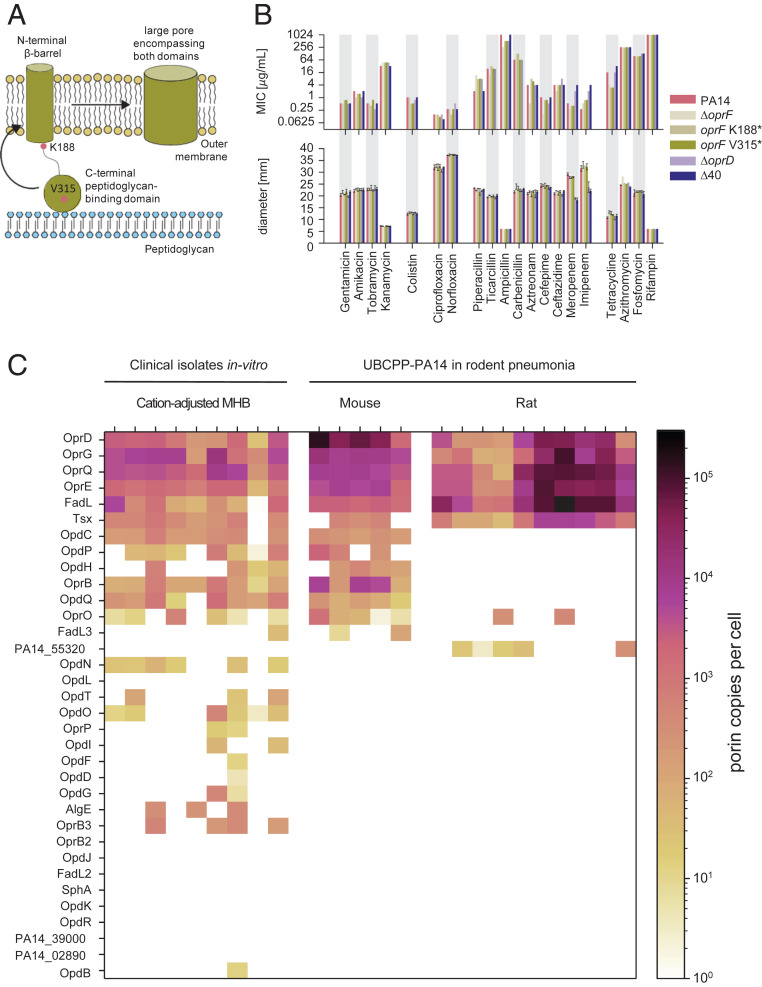
OprF is one of the most abundant proteins in the P. aeruginosa outer membrane (4). Based on experiments with OprF reconstituted in liposomes, a minor open-channel conformer of OprF has been proposed to be the major entry pathway for various hydrophilic molecules including many antibiotics (7) (Fig. 1A). To test this idea in the native context of intact P. aeruginosa cells, we constructed mutants of the virulent clinical isolate P. aeruginosa UCBPP-PA14 (22) that lacked oprF or expressed chromosomal oprF variants with truncated C termini (oprF K188* and oprF V315*; the amino acid numbers include the 24 amino acids of the signal peptide that are cleaved off during maturation) (23). These mutants would be partially (oprF V315*) or completely (oprF K188*) incapable of enlarging the N-terminal eight-strand β-barrel by incorporating further β-strands from the usually globular peptidoglycan-binding C-terminal domain (7). The ΔoprF mutant would also be unable to form dimers with fused β-barrels (24). All three P. aeruginosa mutants had wild-type fitness in rich culture media, as expected (23).
Fig. 1.
Porin involvement in P. aeruginosa antimicrobial susceptibility. (A) Model of OprF with a large majority of two-domain conformer with a narrow outer-membrane β-barrel and a C-terminal domain linking the outer membrane with peptidoglycan and a minority of one-domain conformer with a large pore. (B) MIC (Upper) and antibiograms (Lower) of wild-type P. aeruginosa UCBPP-PA14 and various porin mutants. Means and SD of three experiments are shown. (C) Porin abundance in clinical P. aeruginosa strains in vitro and in UCBPP-PA14 in two rodent pneumonia models as determined by targeted proteomics.
If the large-channel conformer of OprF is indeed the major entry route for antibiotics as proposed, all three mutants should be less susceptible to antimicrobials. However, minimal inhibitory concentrations (MIC) and inhibition zones in disk diffusion tests (antibiograms) for diverse antibiotics (Fig. 1B) showed only minor differences between wild type and mutants that were mostly within the accuracy of the respective assays (twofold for MIC values and 2 mm for inhibition zones). Piperacilin showed fourfold higher MIC against oprF mutants, but this was inconsistent with unaltered inhibition zones. A minor impact of oprF mutations on susceptibility to β-lactam antibiotics has previously been reported (13). Tetracycline had increased activity against oprF mutants in both assays, suggesting potential indirect effects of dysfunctional OprF increasing sensitivity to this translational inhibitor, but not to aminoglycosides. The overall limited impact of OprF on antimicrobial susceptibility was not a result of increased outer-membrane permeability compensating for reduced entry through OprF in the mutants, because azithromycin and rifampin, which are sensitive indicators for outer-membrane barrier function in Pseudomonas (25, 26), remained poorly active in all three mutants. Together, these data indicate a limited role of OprF in antimicrobial translocation across the outer membrane.
“Substrate-Specific” Porins Have Limited Impact on Antimicrobial Translocation.
In addition to OprF, P. aeruginosa encodes dozens of “substrate-specific” porins that might mediate antibiotic uptake (19). One of these porins, OprD (also called OccD1), facilitates translocation of carbapenems (18), and OpdP/OccD3 might also contribute to this under special circumstances (27). In addition, OpdH/OccK5 and OprE/OccK8 have been implicated in translocation of the cephalosporin ceftazidime (28, 29), whereas OprO and OprP can transport fosmidomycin in vitro (30). To determine the relevance of these and other porins for the translocation of diverse antimicrobials in intact bacteria, we constructed a series of porin deletion mutants. We generated clean gene deletions to minimize potential polar effects on the expression of downstream genes. Initial characterization showed unaltered susceptibilities (with the exception of the known OprD–carbapenem link) in agreement with previous “resistome” data (27, 31–34). To test the possibility that phenotypes of single porin mutants were buffered by other porins, we mined the PA14 genome and identified a total of 40 porin candidates (SI Appendix, Table S1). Several combinations of porin deletions still resulted in unaltered antibiotic susceptibilities.
Eventually, we generated a strain, PA14 Δ40, that lacks all 40 porin genes as verified by whole-genome sequencing. During the construction this strain acquired nine secondary mutations including the loss of a duplicate transfer RNA-Asp gene and two nonsynonymous mutations in protein-encoding genes. None of the affected genes had a known association with outer-membrane permeability or antimicrobial susceptibility (32–34) (SI Appendix, Supplementary Information Text and Table S2). PA14 Δ40 grew at rates comparable to the wild type in rich culture media and showed wild-type susceptibility to diverse antimicrobials under standard assay conditions (Fig. 1B). The only clear change was a moderately reduced susceptibility (i.e., higher MIC values and smaller inhibition zones) to the carbapenems meropenem and imipenem, which could be explained almost entirely by the well-known role of OprD/OccD1.
Together, these data demonstrate that the 40 porins are not the major entry pathway for antibiotics under standard conditions. We cannot exclude that induction of certain porins with low expression levels in standard Mueller–Hinton medium might permit antimicrobial entry under nonstandard conditions (27, 28). However, mass spectrometry–based proteome analysis revealed that P. aeruginosa porin abundance in Mueller–Hinton broth closely mimics porin patterns as observed in two different rodent infection models (Fig. 1C), suggesting that standard assays comprise all clinically relevant porins.
Diverging Requirements of “Specific” Porins for Nutrient Uptake.
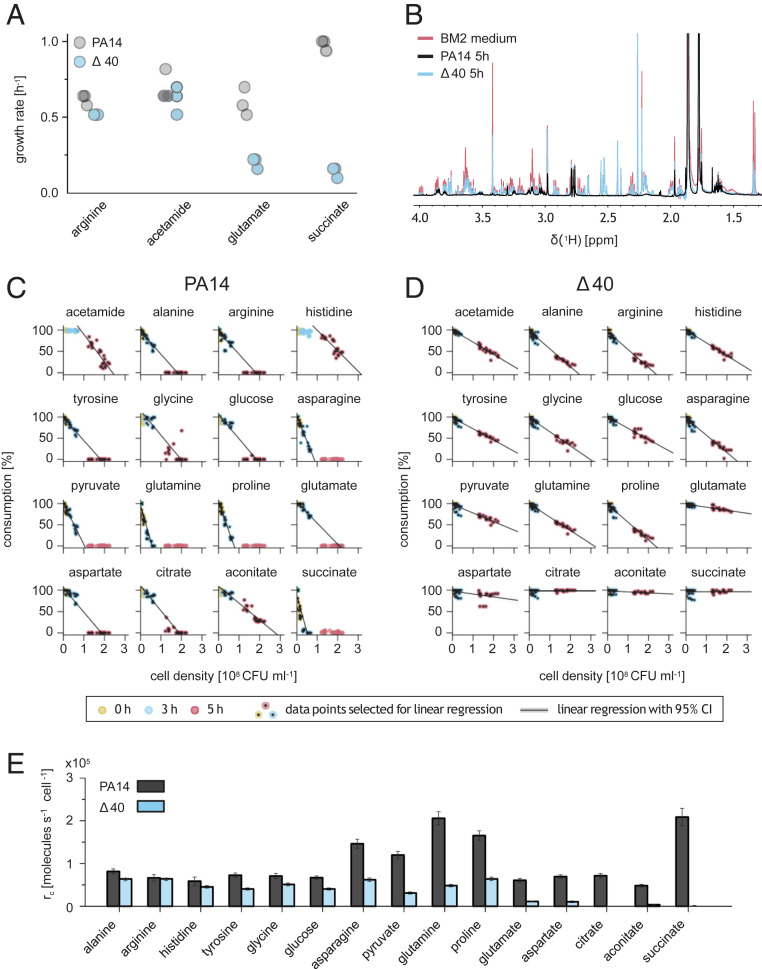
The PA14 Δ40 strain grew normally on rich media and on minimal media containing 10 mM acetamide or arginine as the sole carbon/energy source but poorly on minimal media containing the otherwise preferred carbon/energy sources (35) glutamate or succinate (Fig. 2A and SI Appendix, Fig. S1A), indicating a key role of porins for efficient uptake of some but not all nutrients. To test this idea, we mixed 16 chemically diverse carbon/energy sources (organic acids, amino acids, and glucose) that are known to be utilized by PA14 (36) at concentrations of 100 μM and quantified their consumption by PA14 wild type and PA14 Δ40 using NMR spectroscopy (37) (Fig. 2B and SI Appendix, Figs. S1B and S2). We used comparatively low nutrient concentrations to reduce interference by other potentially rate-limiting steps in nutrient utilization (i.e., transport across the inner membrane and catabolism) (38) and to minimize the contribution of inefficient unspecific translocation pathways (31). To enable consistent growth of PA14 Δ40 (and PA14), we also included 10 mM acetamide, which was readily consumed by both strains (Fig. 2 A, C, and D and SI Appendix, Fig. S1A). We preadapted the strains to this medium to ensure proper induction of respective utilization pathways and to minimize lag phases that occurred after switching media with different nutrients.
Fig. 2.
Porin dependency of P. aeruginosa nutrient uptake. (A) Growth rates of P. aeruginosa PA14 wild-type and porin-free PA14 Δ40 in BM2 minimal media containing a single energy/carbon source. (B) One-dimensional 1H-NMR spectrum of modified BM2 medium containing 16 different nutrients before and after 5 h growth of P. aeruginosa PA14 wild type or porin-free PA14 Δ40. (C and D) PA14 nutrient consumption as measured by one-dimensional [1H] NMR spectroscopy. Each dot represents individual data for 1 of 21 independent cultures. (E) PA14 Δ40 nutrient consumption as measured by one-dimensional [1H] NMR spectroscopy. Each dot represents individual data for 1 of 21 independent cultures. Uptake rates for individual nutrients based on data shown in C and D. Means and SDs are shown.
P. aeruginosa wild type consumed all 16 components within a few hours with the typical pseudomonal preference for succinate, glutamine, proline, and asparagine (35) (Fig. 2 C and D). The porin-free strain PA14 Δ40 consumed small (alanine) and positively charged (histidine and arginine) nutrients at 80 to 100% of the wild-type rates (Fig. 2E). Although some porins such as OprD/OccD1 and closely related paralogs can permit translocation of arginine (20), they were obviously not required for wild-type arginine consumption rates (31). By contrast, all nutrients that had two or more carboxylate groups (aconitate, aspartate, citrate, glutamate, and succinate) showed marked porin dependency (PA14 Δ40 had <20% of wild-type consumption rates). Other compounds had intermediate porin dependency (asparagine, glycine, glucose, glutamine, proline, pyruvate, and tyrosine), indicating that the porin-free outer membrane was partially permeable for these compounds, but porins facilitated translocation.
Together, these data showed that the outer membrane of P. aeruginosa permitted entry of hydrophilic/amphiphilic compounds with substantial rates, even in the absence of all 40 identifiable porins. This includes the preferred carbon sources alanine and arginine but also other amino acids and glucose that are accessible for P. aeruginosa in millimolar concentrations in the lung of cystic fibrosis (CF) and non-CF patients (39, 40). This compound uptake was not caused by general membrane leakage/permeabilization in the absence of porins because small molecules with two or more carboxylate groups (such as succinate) were effectively excluded.
Complex Substrate Selectivity of Individual Porins.
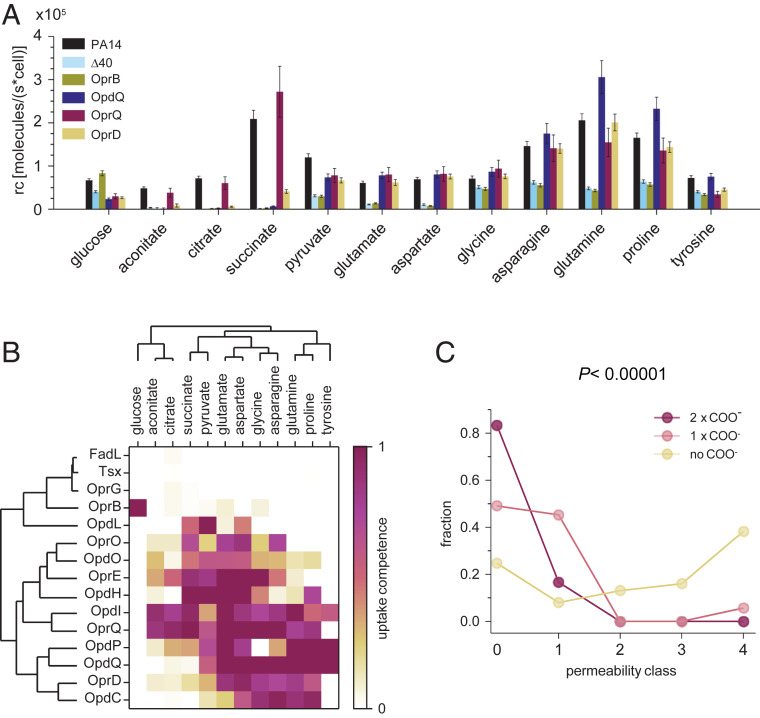
To determine the contribution of individual porins to nutrient uptake, we expressed single porin genes in the porin-free PA14 Δ40 background. For all constructs we used the same PoprD promoter on low-copy plasmids to minimize interference by specific porin-induction patterns (28). This strategy enabled determination of nutrient translocation through a single porin in the native membrane context without interference by other porins. We focused on 13 porins that we detected in various rodent infection models and in diverse clinical isolates grown under standard conditions for clinical microbiology (Mueller–Hinton broth) (Fig. 1C). This set included all nine porins detected in the lung of CF patients (41). For comparison, we included also the two porins OpdI/OccD5 and OpdL/OccK4 that were poorly expressed under all these conditions (“cryptic” porins).
FadL and Tsx had no impact on consumption of any of the tested nutrients (Fig. 3 A and B and SI Appendix, Fig. S3), consistent with their proposed selectivity for fatty acids and nucleosides, respectively, which were not included in our nutrient mix. OprG had no detectable impact on nutrient consumption, arguing against an important role for uptake of small amino acids including glycine and alanine (42) under native conditions, consistent with its narrow channel consisting of eight β-strands (19). OprB enabled wild-type consumption rates specifically for glucose, as expected (38). The remaining porins enabled consumption of 3 to 11 different nutrients at near wild-type levels. Importantly, no porin except for OprB enabled consumption of glucose, indicating no general membrane leakage as a result of forced porin expression.
Fig. 3.
Nutrient uptake through single porins in absence of other porins. (A) Nutrient consumption rates of PA14 Δ40 strains expressing a single porin from a low-copy-number plasmid. Means and averages for three independent cultures (single-porin strains) or 21 cultures (PA14, PA14 Δ40) are shown. (B) Substrate selectivity of 15 porins determined in single-porin strains. The capacity of each porin to boost nutrient consumption from baseline levels in PA14 Δ40 to wild-type levels is shown. Porins and nutrients are grouped based on unsupervised clustering. (C) Permeability of 472 antimicrobial compounds with detectable antipseudomonal activity (15). Following the published analysis, compounds are grouped in permeability classes according to the MIC values for efflux-deficient P. aeruginosa strains with intact (P Δ6) or abolished (P Δ6-pore) outer membrane permeability barrier (class 0, MIC of PA Δ6-pore is less than 20% of PA Δ6; 1, 20 to 40%; 2, 40 to 60%, 3, 60 to 80%; 4 more than 80%). Statistical significance was analyzed with the χ2 test.
Each of the other porins had a distinct substrate spectrum, while at the same time each nutrient was able to translocate through several different porins. The tricarboxylates, citrate and aconitate, entered almost exclusively through just two porins, OpdI/OccD5 and OprQ/OccD6, but hardly through the previously postulated OpdH/OccK5, consistent with negative electrophysiology data (28). The aromatic amino acid tyrosine translocated mainly through OpdP/OccD3 and OpdQ/OccK6, and partially through OpdI/OccD5. The previously implicated OpdT/OccD4 was not among our set of expressed porins. Glutamate translocated efficiently through several porins of both the OpdK/OccK and OprD/OccD families (OprE/OccK8, OprQ/OccD6, OpdI/OccD5, OpdQ/OccK6, and OpdH/OccK5) and partially through OprD/OccD1 and OprO. Results for OprE/OccK8 were consistent with recent liposome swelling assays (29). Succinate also translocated through members of both porin subfamilies (OpdH/OccK5, OpdI/OccD5, OprE/OccK8, and OprQ/OccD6). The “cryptic” porin OpdI/OccD5 had transport capabilities similar to the abundantly expressed and distantly related subfamily member OprQ/OccD6, which might explain why OpdI/OccD5 was not expressed under standard conditions. The “cryptic” porin OpdL/OccK4 had an unusually narrow substrate spectrum with a preference for pyruvate, suggesting that it might be induced when this nutrient is available. The broad range of substrates of some porins contrasted with efficient exclusion of only narrow sets of nutrients (e.g., glycine–OpdP, succinate–OpdQ, and proline–OprE).
These data do not support the previously proposed distinct substrate spectra of porin subfamilies (OprD/OccD1, positively charged amino acids; OpdK/OccK subfamily, net negative charge). This proposal was based on observations from porins expressed in E. coli (20) which might have affected their channel properties. Recent electrophysiology data for OprE/OccK8 also question a simple dichotomy for substrates between the two subfamilies (29). Unsupervised clustering for transport capabilities observed in this study yielded incomplete separation of the two subfamilies and no signature substrates for either subfamily (Fig. 3B). Furthermore, the clustering did not conform with overall sequence similarity within each subfamily (19) (e.g., the sequence of OccD1 is closer to OccD3 and OccD6 than to OccD2, which is, however, more similar in terms of substrates).
Finally, chemically similar substrates did cluster together (tricarboxylates citrate/aconitate; small negatively charged acids succinate/pyruvate; negatively charged amino acids glutamate/aspartate) although the zwitterionic amino acids formed two separate clusters tyrosine/glutamine/proline and glycine/asparagine, possibly driven by molecular size. Together, these data show that certain porins share substrate preferences independently of their evolutionary relatedness. This might facilitate identification of relevant structure–function relationships against commonly inherited channel properties in future studies.
Taken together, these data show 1) substantial porin-independent translocation of certain nutrients and 2) broad porin substrate spectra and marked overlap, but also efficient porin-specific nutrient exclusion.
Carboxylate Groups Slow Translocation.
Porin-independent translocation was crucial for antibiotic activity (Fig. 1 B and C). Compound properties that interfere with this pathway should thus be avoided in antimicrobial discovery and development. Our nutrient consumption data for compounds with masses below 200 Da indicated that two or more carboxylate groups blocked porin-independent translocation (Fig. 2 C and D; aconitate, citrate, and succinate). To extend these observations, we reanalyzed recent datasets for more than 500 antipseudomonal compounds in the 250- to 700-Da size range (14, 15). In particular, we compared MIC values for efflux-deficient P. aeruginosa strains with intact outer membrane barrier (P Δ6, lacking six major efflux systems) or abolished barrier function because of insertion of water-filled pores with ∼2.4-nm diameter (P Δ6-pore). We focused on these efflux-deficient strains to unravel the contribution of the outer membrane translocation with minimal interference by efflux (16). Antimicrobials such as gentamicin do not gain activity in P Δ6-pore compared to P Δ6, suggesting that they can permeate already the wild-type outer membrane efficiently. Large antimicrobials such as azithromycin (758.88 Da) and erythromycin (728.38 Da) are 128-fold more potent against P Δ6-pore compared to P Δ6, indicating that the intact outer membrane provides an efficient barrier for these molecules.
Previous analysis of these data has revealed molecular fragments promoting translocation through the wild-type outer membrane (primary and secondary amine groups, benzene rings, and trifluoromethyl groups) (15). Our reanalysis showed an additional marked negative impact of carboxylate groups on membrane permeation (Fig. 3C). Fifty out of 53 molecules (94%) with one carboxylate group and all six compounds with two carboxylates were bad permeators [“class 0,” activity gain in P Δ6-pore greater than fivefold; “class 1,” gain between 5- and 2.5-fold (15)], while only 135 out of 413 molecules (33%) without carboxylates had such bad permeation capabilities. These results indicate that carboxylate groups slow outer-membrane permeation. This carboxylate penalty supports and extends our findings for porin-independent nutrient uptake (Fig. 2 C and D).
It is important to note that the antimicrobial ceftazidime has two carboxylate groups and is a “poor permeator” [64-fold activity gain in P Δ6-pore (14)], yet it remains the first choice for treating patients with susceptible P. aeruginosa infections (43). Poor permeation is thus compatible with clinical efficacy if other aspects such as target binding and efflux avoidance are favorable (as is the case for ceftazidime). Efficient translocation is, however, crucial for compounds with less-favorable efflux and target-binding properties (4, 16, 44).
Discussion
Porins with water-filled channels are commonly assumed to be essential for efficient translocation of hydrophilic compounds across the outer membrane of Gram-negative bacteria. Our comprehensive deletion of all porin genes in a clinically relevant P. aeruginosa isolate and the generation of a series of P. aeruginosa strains that carry only one type of porin provided unique opportunities for determining porin function in the native outer-membrane environment.
We observed that the porin-free mutant PA14 Δ40 grew normally on rich media and, unexpectedly, also on minimal media containing a single, hydrophilic carbon/energy source. Quantification of consumption rates using NMR spectroscopy showed porin-independent uptake of diverse water-soluble molecules with rates in the range of 50,000 molecules s−1 per cell at an external concentration of 100 μM. However, nutrients with two or more carboxylate groups permeated poorly in the absence of porins. These results indicate a remarkable permeability of the porin-free outer membrane for many but not all hydrophilic molecules.
In addition to porins, the Bam complex (45, 46), secretins (47), and outer-membrane channels of efflux systems (3) are thought to transiently form large, unspecific pores in the outer membrane. However, such large unspecific pores seem to play a limited role for nutrient uptake by P. aeruginosa because small molecules such as succinate and citrate permeate only slowly in the absence of porins. Instead, many molecules likely translocate directly through the lipid bilayer (15, 48–52). This translocation pathway is consistent with the preference for amines and effective exclusion of carboxylates: Amines can efficiently compete with Mg2+ and Ca2+ cations (53) that form salt bridges with phosphate and carboxylate groups of LPS, thereby weakening the gel-like Mg2+/phosphate/LPS core clusters and enhancing translocation [the “self-promoted uptake” concept (48)]. By contrast, carboxylates might displace water molecules of the inner hydration shell of Mg2+, thus forming stable complexes (54) that link the carboxylates to the rigid gel-like Mg2+/phosphate/LPS core clusters without weakening them, thereby slowing permeation.
Although the outer-membrane lipid bilayer enabled substantial porin-independent permeation by many hydrophilic molecules, valuable and preferred nutrients with two or more carboxylate groups such as succinate had low permeation rates in the absence of porins. The large OprD/Occ family of porins may have evolved specifically for uptake of compounds with carboxylate groups (20), thus opening translocation pathways for these poorly permeating but valuable nutrients. Many porins had unexpectedly broad substrate spectra, which challenges the one porin–one nutrient model. On the other hand, each porin excluded effectively individual nutrients with high specificity, although these nutrients are readily transported by closely related porin family members. This combination of promiscuous uptake and highly specific exclusion suggests that porin evolution was mostly shaped by selection against the entry of toxic compounds while specific uptake of only one nutrient was not critical. Most nutrients that could use various porins for entry were covered by a combination of just two porins, OprE/OccK8 and OprQ/OccD6, which are abundant in various P. aeruginosa clinical isolates when grown in standard medium, and in PA14 in different rodent models (Fig. 1C). Uptake of additional substrates could be covered by substrate-induced expression of other porins (31). This substrate overlap may also explain why clinical isolates often down-regulate or inactivate OprD/OccD1, resulting in diminished susceptibility to carbapenems (55). This loss should not impair Pseudomonas nutrition because the abundant porins OprE/OccK8 and OprQ/OccD6 cover the same nutrients as OprD/OccD1 (at least among the nutrients tested in this study). Indeed, OprD loss in clinical isolates has no detectable fitness costs in rodent pneumonia models (in fact, fitness is greatly increased for unknown reasons) (56). Porin redundancy thus facilitates emergence of carbapenem resistance.
Our antimicrobial susceptibility data show that neither the “general” porin OprF nor the “specific” porins were relevant for translocation of diverse antimicrobials under standard conditions, with the sole exception of previously identified partial permeation of carbapenems through OprD. These results provide further support for the relevance of porin-independent permeation of the outer-membrane lipid bilayer. Recent data confirm that E. coli lacking both major unspecific porins OmpC and OmpF show decreased susceptibility to several β-lactam antibiotics (9), consistent with the role of the major porins for uptake of these drugs (13). However, some β-lactams and diverse other antibiotics retain most of their activity against this ompC ompF mutant (9), suggesting efficient alternative entry pathways. This could suggest a similar porin-independent entry pathway, but other still-remaining porins or disturbed barrier function in the mutant could also be involved.
In conclusion, diverse hydrophilic compounds can penetrate the P. aeruginosa outer-membrane bilayer at relevant rates independently of porins. Porins are mostly required for utilization of valuable nutrients containing multiple carboxylate groups, which permeate poorly through the porin-free outer membrane. Porins provide efficient translocation pathways for these nutrients but efficiently exclude antibiotics. Antibiotics thus have to enter the cell by direct penetration of the outer-membrane lipid bilayer, resulting in a penalty for molecules carrying carboxylate groups. Thus, replacement of carboxylate groups (which are present in many current antibiotics) by isosteres (57) might be considered to accelerate compound translocation across the P. aeruginosa outer membrane. Future studies should further characterize this largely neglected translocation pathway, to identify additional molecular properties that determine translocation rates of antimicrobials.
Materials and Methods
Bacterial Strains and Growth Conditions.
All P. aeruginosa mutants used in this study are derived from the clinical isolate UCBBP-PA14 (22). In addition, we analyzed various P. aeruginosa clinical isolates from the University Hospital Basel strain collection. E. coli Sm10λpir was used for cloning and to conjugate plasmids into P. aeruginosa. All bacteria were cultured at 37 °C in lysogeny broth (LB) except for mating, for which we used P. aeruginosa grown overnight at 42 °C. For growth assays, bacteria were grown in overnight in LB and then overnight in basal medium 2 (BM2; http://cmdr.ubc.ca/bobh/method/media-recipes/) minimal medium with 10 mM acetamide as carbon source. After washing, the bacterial were inoculated in BM2 containing the carbon source of choice at an initial optical density at 600 nm (OD600) = 0.05.
Antibiotics and Reagents.
Amikacin (disulfate salt; potency 77.60%), aztreonam (potency 92%), azithromycin (potency 92.70%), cefepime (hydrochloride, 83.82%), ciprofloxacin (potency 78.60%), colistin (sulfate salt; potency, 67.50%), gentamicin (sulfate salt, potency 67.70%), imipenem (monohydrate, potency 93.66%), kanamycin (sulfate salt, potency 83.16%), meropenem (trihydrate, potency 87.64%), piperacillin (sodium salt, potency 94.60%), tetracycline (potency 100%), ticarcillin (disodium salt, potency 90.62%), and tobramycin (potency 95.20%) were purchased from Sigma-Aldrich. Ampicillin (disulfate salt, potency 77.60%) and carbenicillin (disodium salt, potency 89.58%) were purchased from Roth. Ceftazidime was purchased from European Pharmacopeia. Unless stated otherwise, all other reagents were of analytical grade and were purchased from Sigma-Aldrich-Fluka.
Gene Deletion and Episomal Porin Expression.
Strains and plasmids used in this study are listed in SI Appendix, Table S3. Primers are listed in SI Appendix, Table S4.
Knockout vectors were constructed as described (58) with the following modifications. Seven hundred-base pair sequences of the flanking regions of the porin gene were PCR-amplified with primers designed with Snapgene software (GSL Biotech LLC). The fragments were gel-purified and inserted into pEXG2 plasmid (59) by Gibson assembly (60). The assembled plasmid was transformed into competent SM10λpir prepared with Mix & Go (Zymo Research Corporation) and plated on LB agar containing 50 µg/mL kanamycin and 15 µg/mL gentamicin. Sequenced-verified clones were mated for 4 h with PA14 strains at 37 °C. Single cross-over events were selected on plates containing 15 µg/mL gentamicin and 20 µg/mL irgasan. Colonies were picked and grown in LB for 4 h and streaked on 5% sucrose plates overnight at 30 °C. P. aeruginosa clones were confirmed by sequencing and stored at −80 °C in LB containing 9% dimethyl sulfoxide. Whole-genome sequencing was done as described previously (58). Geneious Prime 2019.0.4 was used to map the reads to the P. aeruginosa UCBPP-PA14 reference sequence NC_008463.1 and to identify variations.
A plasmid backbone for expression of individual porins was constructed using Gibson assembly of the TrfA-OriV origin of replication from pAD6 (61), a derivative of the low-copy-number plasmid RK2 (62), the gentamicin resistance cassette and origin of transfer (oriT) from pEXG2, rpsI and rrnB terminators, and the PoprD promoter amplified from PA14. A porin gene was inserted downstream of PoprD. For electroporation of porin-expression plasmids, 20-mL PA14 Δ40 overnight cultures in LB were washed thrice with ice-cold 0.3 M sucrose and resuspended in 100 µL cold 0.3 M sucrose. Electroporation was done with 1 μL plasmid solution in 2-mm cuvettes at 25 µF/400 Ohm/2.5 kV. After addition of 1 mL prewarmed LB and incubation for 1 h at 37 °C, cells were plated on LB agar containing 15 µg/mL gentamicin.
Drug Susceptibility Tests.
The MIC of drugs was determined by a twofold dilution assay in a 96-well plate according to Clinical and Laboratory Standards Institute guidelines (inoculum of ∼106 colony-forming units [CFU]/mL; reading after 20- to 24-h incubation) in cation-adjusted Mueller–Hinton broth (63). Growth of bacteria at 37 °C was examined by visual inspection after 20-h incubation. The MIC was defined as the lowest concentration of an antibiotic that completely prevented visible cell growth. Drug susceptibility was also determined with antibiogram measurements with 20 different antibiotics (Bio-Rad commercial disk). Overnight cultures of P. aeruginosa strains were diluted at OD600 = 0.1 and were spread on 120- × 120-mm2 MHB II plates and air-dried in a laminar flow and then discs containing antibiotics were placed on the plates. Plates were incubated at 37 °C for 20 h. The diameter of halos surrounding the discs were measured as an indication of growth inhibition.
Nutrient Consumption Assays.
P. aeruginosa strains were grown overnight in defined nutrient medium (BM2 containing 10 mM acetamide and 100 μM of 10 amino acids [alanine, arginine, asparagine, aspartate, glutamate, glutamine, glycine, histidine, proline, tyrosine], glucose, cis-aconitate, citrate, succinate, and pyruvate). Bacteria were washed and resuspended in prewarmed nutrient medium at OD600 = 0.005. Cultures were incubated at 37 °C and 180 rpm and samples were taken after 2, 3, and 5 h of growth. OD600 and CFU were determined at each time point. The remaining volumes were filtered through a 0.2-μm pore filter and stored at −80 °C until NMR analysis.
NMR spectra were measured on a 600-MHz Bruker Avance III HD NMR spectrometer equipped with a cryogenic QCI-F probe. One-dimensional [1H] spectra were recorded with a free induction decay size of 32,000 points and 256 transients at 298 K. Water was suppressed by excitation sculpting. Spectra were processed using TopSpin 3.6 by applying an exponential window function with line broadening factor of 0.3 Hz and zero filling to 64,000 points prior to Fourier transformation. For each substance, an isolated signal was chosen for analysis (SI Appendix, Fig. S2). Peak intensities were determined by comparison with nutrient medium as reference.
Analysis of Consumption Kinetics.
We assumed that during exponential growth, rC, the average consumption rate per bacterium of nutrient X from the medium is constant. The consumption rate of nutrient X by the bacterial population is thus proportional to the cell density n(t):
where n0 is the density of bacteria at t = 0 and k the growth rate constant. Integration results in the residual substance concentration at time T:
and the total nutrient consumption MX, which depends linearly on the cell density n(T):
The cell density relates to the OD600 via a proportionality factor :
where N is the number of CFU determined in a volume of cell culture. In separate calibration experiments, was determined to be 9.8 × 108 CFU⋅mL−1 and k was determined for each strain by exponential fitting of growth curves. The consumption rates rC was then obtained from linear fits of data pairs [Mx, n(T)].
To account for differential consumption preferences (delayed uptake characterized by 1) a lag-phase, 2) moderate uptake, and 3) fast uptake), different subsets of data points were used for linear regression modeling (as indicated in Fig. 1C):
-
1)
Data points at t = 3 and 5 h
-
2)
Data points at t = 0, 2, 3, and 5 h
-
3)
Data points at t = 0, 2 and 3 h.
For calculating the uptake competence of each porin for each nutrient X, the consumption rate of the single-porin strain for X was normalized to a range defined by the value of the consumption rates of PA14 and Δ40:
Values below 0 were set to 0 and values above 100 were set to 100.
Proteomics.
P. aeruginosa porins were detected by targeted proteomics using parallel reaction monitoring on a high-resolution and accurate mass instrument with absolute quantification using heavy-isotope-labeled reference peptides as described previously (29). We analyzed PA14 and various clinical isolates grown to exponential phase in cation-adjusted Mueller–Hinton broth. We also reanalyzed previously obtained blood or lung homogenates from mice and rats (29) that had been obtained at 24 h postinfection by intratracheal instillation of an agar bead containing 107 CFU of PA14.
Curve Fittings and Statistical Analysis.
Curve fitting analyses and calculations of regression parameters were made with GraphPad Prism (version 4.03) software for Windows and statistical analysis with GraphPad Instat version 3.06 for Windows.
Supplementary Material
Acknowledgments
The work was supported by the European Union and the European Federation of Pharmaceutical Industries and Associations (IMI, ND4BB-TRANSLOCATION) and Swiss National Science Foundation (310030_156818, 310030_182315, NRP 72-177449, NCCR AntiResist) to D.B. and Biozentrum PhD fellowships to J.U. and V.T. We thank Prof. Dr. Daniel Häussinger for providing access to his NMR spectrometers and fruitful discussions on data analysis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107644118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Tacconelli E., et al., Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Hancock R. E., The bacterial outer membrane as a drug barrier. Trends Microbiol. 5, 37–42 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Zgurskaya H. I., Löpez C. A., Gnanakaran S., Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1, 512–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vergalli J., et al., Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Snyder D. S., McIntosh T. J., The lipopolysaccharide barrier: Correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry 39, 11777–11787 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Sugawara E., Nagano K., Nikaido H., Alternative folding pathways of the major porin OprF of Pseudomonas aeruginosa. FEBS J. 279, 910–918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delcour A. H., Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794, 808–816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi U., Lee C. R., Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 10, 953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter M. F., et al., Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 545, 299–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acosta-Gutiérrez S., et al., Getting drugs into Gram-negative bacteria: Rational rules for permeation through general porins. ACS Infect. Dis. 4, 1487–1498 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Parker E. N., et al., Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat. Microbiol. 5, 67–75 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock R. E., Woodruff W. A., Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev. Infect. Dis. 10, 770–775 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Cooper C. J., et al., Molecular properties that define the activities of antibiotics in Escherichia coli and Pseudomonas aeruginosa. ACS Infect. Dis. 4, 1223–1234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansbach R. A., et al., Machine learning algorithm identifies an antibiotic vocabulary for permeating Gram-negative bacteria. J. Chem. Inf. Model. 60, 2838–2847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zgurskaya H. I., Rybenkov V. V., Permeability barriers of Gram-negative pathogens. Ann. N. Y. Acad. Sci. 1459, 5–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tommasi R., Brown D. G., Walkup G. K., Manchester J. I., Miller A. A., ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14, 529–542 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Quinn J. P., Dudek E. J., DiVincenzo C. A., Lucks D. A., Lerner S. A., Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J. Infect. Dis. 154, 289–294 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Chevalier S., et al., Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41, 698–722 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Eren E., et al., Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biol. 10, e1001242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dogan Guzel F., et al., Towards understanding single-channel characteristics of OccK8 purified from Pseudomonas aeruginosa. Eur. Biophys. J. 50, 87–98 (2021). [DOI] [PubMed] [Google Scholar]
- 22.He J., et al., The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. U.S.A. 101, 2530–2535 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawling E. G., Brinkman F. S., Hancock R. E., Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180, 3556–3562 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishida H., Garcia-Herrero A., Vogel H. J., The periplasmic domain of Escherichia coli outer membrane protein A can undergo a localized temperature dependent structural transition. Biochim. Biophys. Acta 1838, 3014–3024 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Balibar C. J., Grabowicz M., Mutant alleles of lptD increase the permeability of Pseudomonas aeruginosa and define determinants of intrinsic resistance to antibiotics. Antimicrob. Agents Chemother. 60, 845–854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnamoorthy G., et al., Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. MBio 8, e01172-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isabella V. M., et al., Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa. Chem. Biol. 22, 535–547 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Tamber S., Maier E., Benz R., Hancock R. E., Characterization of OpdH, a Pseudomonas aeruginosa porin involved in the uptake of tricarboxylates. J. Bacteriol. 189, 929–939 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samanta S., et al., Getting drugs through small pores: Exploiting the porins pathway in Pseudomonas aeruginosa. ACS Infect. Dis. 4, 1519–1528 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Piselli C., Benz R., Fosmidomycin transport through the phosphate-specific porins OprO and OprP of Pseudomonas aeruginosa. Mol. Microbiol., 10.1111/mmi.14693 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Tamber S., Ochs M. M., Hancock R. E., Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J. Bacteriol. 188, 45–54 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breidenstein E. B., Khaira B. K., Wiegand I., Overhage J., Hancock R. E., Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52, 4486–4491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dötsch A., et al., Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 2522–2531 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E., Martínez J. L., Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob. Agents Chemother. 54, 4159–4167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojo F., Carbon catabolite repression in Pseudomonas: Optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34, 658–684 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Bartell J. A., et al., Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat. Commun. 8, 14631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behrends V., Williams H. D., Bundy J. G., Metabolic footprinting: Extracellular metabolomic analysis. Methods Mol. Biol. 1149, 281–292 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Wylie J. L., Worobec E. A., The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177, 3021–3026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer K. L., Aye L. M., Whiteley M., Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Rosa R., Johansen H. K., Molin S., Adapting to the airways: Metabolic requirements of Pseudomonas aeruginosa during the infection of cystic fibrosis patients. Metabolites 9, E234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X., et al., In vivo proteome of Pseudomonas aeruginosa in airways of cystic fibrosis patients. J. Proteome Res. 18, 2601–2612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucharska I., Seelheim P., Edrington T., Liang B., Tamm L. K., OprG harnesses the dynamics of its extracellular loops to transport small amino acids across the outer membrane of Pseudomonas aeruginosa. Structure 23, 2234–2245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babich T., et al., Ceftazidime, carbapenems, or piperacillin-tazobactam as single definitive therapy for Pseudomonas aeruginosa bloodstream infection: A multisite retrospective study. Clin. Infect. Dis. 70, 2270–2280 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Nichols W. W., “Permeability of bacteria to antibacterial agents” in Antibiotic Discovery and Development, Dougherty T., Pucci M., Eds. (Springer, 2012), pp. 849–879. [Google Scholar]
- 45.Robert V., et al., Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estrada Mallarino L., et al., TtOmp85, a β-barrel assembly protein, functions by barrel augmentation. Biochemistry 54, 844–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Disconzi E., et al., Bacterial secretins form constitutively open pores akin to general porins. J. Bacteriol. 196, 121–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock R. E., Raffle V. J., Nicas T. I., Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 19, 777–785 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitahara T., Yoneyama H., Nakae T., Antibiotic diffusion pathways in the outer membrane of Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 238, 457–461 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Plesiat P., Aires J. R., Godard C., Köhler T., Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 179, 7004–7010 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.López C. A., Zgurskaya H., Gnanakaran S., Molecular characterization of the outer membrane of Pseudomonas aeruginosa. Biochim. Biophys. Acta Biomembr. 1862, 183151 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Patel D. S., et al., Dynamics and interactions of OmpF and LPS: Influence on pore accessibility and ion permeability. Biophys. J. 110, 930–938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galanos C., Lüderitz O., Electrodialysis of lipopolysaccharides and their conversion to uniform salt forms. Eur. J. Biochem. 54, 603–610 (1975). [DOI] [PubMed] [Google Scholar]
- 54.Dudev T., Cowan J., Lim C., Competitive binding in magnesium coordination chemistry: Water versus ligands of biological interest. J. Am. Chem. Soc. 121, 7665–7673 (1999). [Google Scholar]
- 55.Lister P. D., Wolter D. J., Hanson N. D., Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roux D., et al., Fitness cost of antibiotic susceptibility during bacterial infection. Sci. Transl. Med. 7, 297ra114 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Ballatore C., Huryn D. M., A. B. Smith, 3rd, Carboxylic acid (bio)isosteres in drug design. ChemMedChem 8, 385–395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunrath O., et al., Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine 41, 479–487 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rietsch A., Vallet-Gely I., Dove S. L., Mekalanos J. J., ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson D. G., et al., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Malone J. G., et al., YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6, e1000804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jahn M., Vorpahl C., Hübschmann T., Harms H., Müller S., Copy number variability of expression plasmids determined by cell sorting and Droplet Digital PCR. Microb. Cell Fact. 15, 211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clinical and Laboratory Standards Institute, “Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically” (CLSI document M7-A7, Clinical and Laboratory Standards Institute, Wayne, PA, 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.