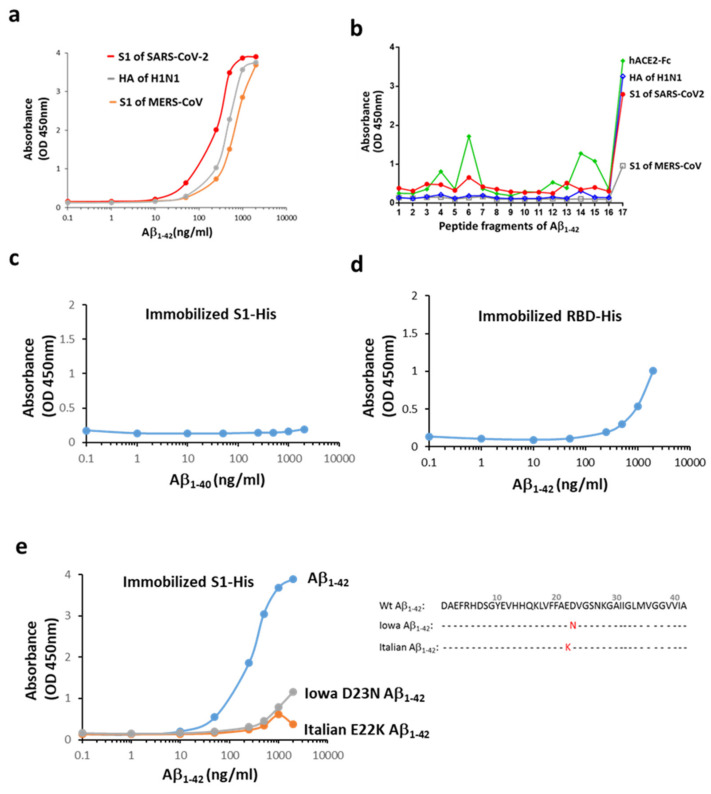
Figure 1.
Interactions between Aβ1-42 and the S1 of SARS-CoV-2. (a) Aβ1-42 differentially binds to the immobilized surface proteins of three viruses responsible for recent pandemics. The order of potency of binding based on the estimated EC50 was the S1 of SARS-CoV-2 (200–325 ng/mL) > HA of H1N1 (HA, 372–507 ng/mL) > S1 of MERS-CoV (599–860 ng/mL). (b) Linear epitope mapping was performed using 17 fragment peptides of Aβ1-42 (details in Supplementary Figure S2), and the binding ability is indicated by OD. Aβ1-42 interacted with the S1 of SARS-CoV-2, mainly in the last probe containing the hydrophobic Aβ residues of 33–42. A similar binding pattern on Aβ1-42 was observed for HA of H1N1, and to a lesser extent, for the S1 of MERS-CoV. hACE2-Fc presents three more binding sites (OD > 0.8) across the entire sequence of Aβ1-42 in addition to a major binding site on the C-terminal end. The following binding potency was compared by OD. (c) Aβ1-40 did not bind to the S1 of SARS-CoV-2. (d) Aβ1-42 bound weakly to the immobilized His-tagged RBD of SARS-CoV-2 as compared to the S1 protein. (e) Iowa D23N and Italian E22K Aβ1-42 mutants exerted marked reduction in the S1 binding, and amino acid sequences for normal human (Wt) and mutated Aβ1-42 are presented. Data are presented as mean values from two independent experiments.