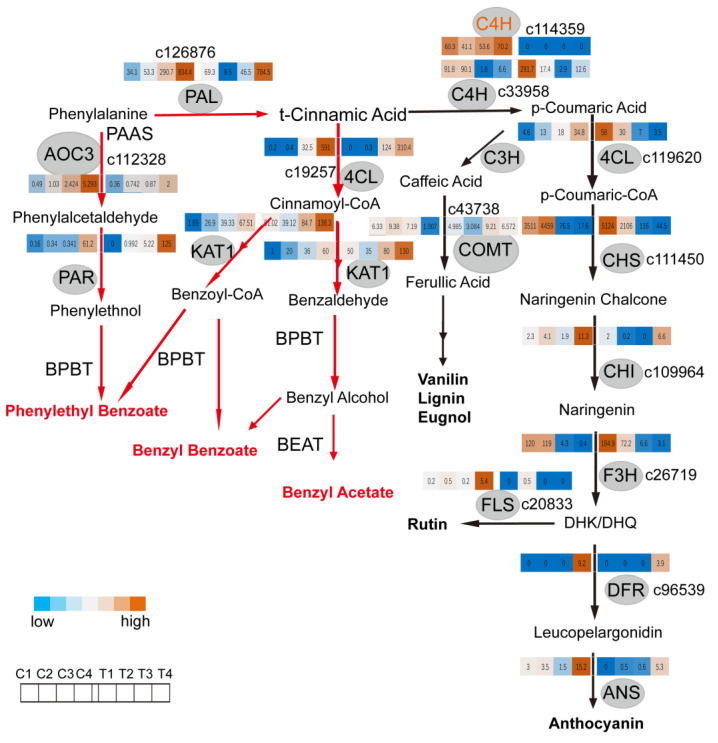
Figure 4.
Expression patterns of DEGs related to branches of the phenylpropanoid pathway in tepals and coronas of Narcissus during flower development. Branches of the phenylpropanoid pathway in Narcissus and expression patterns analysis of DEGs, are related to flavonoids metabolic pathway and benzenoid/phenylpropanoid metabolic pathway. The red line and arrowhead in the left panel represent the phenylpropanoid biosynthesis pathway, and the black line and arrowhead represent the flavonoid/anthocyanin biosynthesis pathway. The heat map shows the expression patterns of differentially expressed structural genes related to branches of the phenylpropanoid pathway in the tepals and coronas at different periods (left down). Each row means one unigene and the expression changes of unigenes are shown by color grids based on FPKM value (fragments per kilobase of exon per million reads mapped). The color bar from blue to orange shows upregulated; from orange to blue is downregulated. Enzyme names are shown in the gray background circle. 4CL, 4-coumarate CoA ligase; KAT, 3-ketoacyl-CoA thiolase; C4H, cinnamic acid 4-hydroxylase; AOC3, amine oxidase, copper containing 3; PAR, phenylacetaldehyde reductase; BEAT, acetyl-CoA: benzylalcohol acetyltransferase; BPBT, benzoyl-CoA: benzoyl-CoA: benzyl alcohol/phenylethanol benzoyltransferas; PAL, phenylalanine ammonia-lyase; PAAS, phenylacetaldehyde synthase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, favanone 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanin synthase; C3H, p-coumarate 3-hydroxylase; COMT, caffeic acid 3-o-methyltransferase.