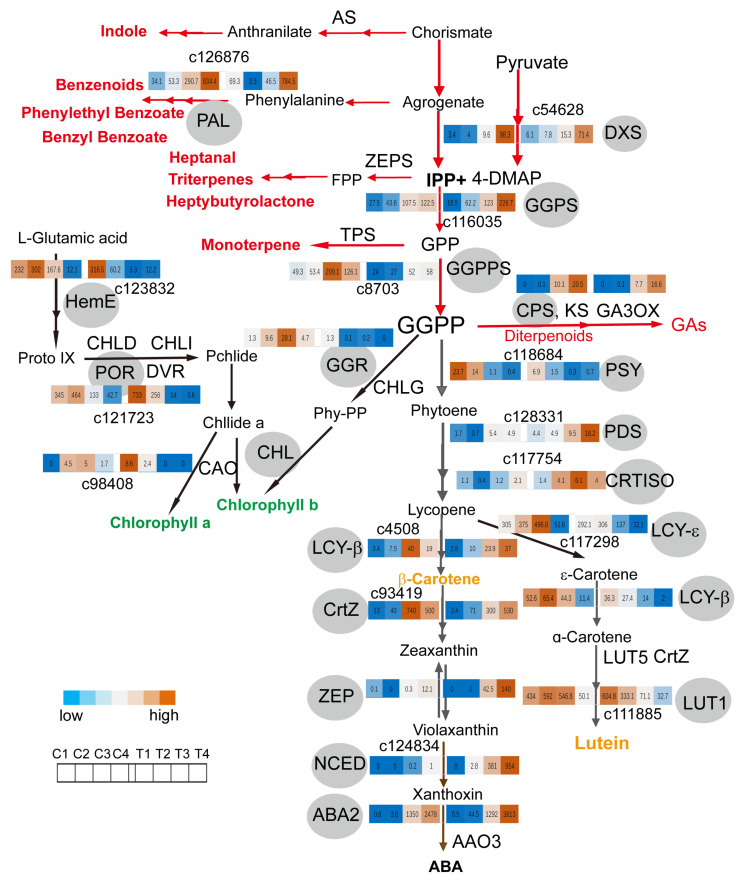
Figure 5.
Three branches of MEP pathway include carotenoids biosynthesis, the biosynthesis of the phytol side chain of chlorophylls, and of diterpenoids such as gibberellins in the Narcissus flower. The red line and arrowhead in the up panel represents the terpenoids metabolic pathway, and the black line and arrowhead represents carotenoids biosynthesis; the biosynthesis of the phytol side chain of the chlorophylls pathway. The heat map shows the expression patterns of differentially expressed genes related to three branches of MEP pathway in tepals and coronas at different periods (left down). Enzyme names are shown in the gray background circle. The FPKM values of genes are labeled on the heat map. The color bar from blue to orange is upregulated; from orange to blue is down regulated: AS, anthranilate synthase; ZFPS, cis,cis-farnesyl diphosphate synthase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; GGPP, geranylgeranyl diphosphate; GGPPS, geranyl diphosphate synthase; TPS, terpene synthase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, zeta-carotene desaturase; CRTISO, carotene isomerase; LCY-ε, lycopene epsilon-cyclase; LCY-β, lycopene beta-cyclase; CrtZ, beta-carotene 3-hydroxylase; LUT5, beta-ring hydroxylase; LUT1, carotenoid epsilon hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; ABA2, xanthoxin dehydrogenase; AAO3, abscisic-aldehyde oxidase; GGR, geranylgeranyl reductase; HemL, glutamate-1-semialdehyde 2,1-aminomutase; HemE, uroporphyrinogen decarboxylase; ChlD, magnesium chelatase subunit D; CHLI, magnesium chelatase subunit I; POR, protochlorophyllide reductase; CAO, chlorophyllide a oxygenase; CLH, chlorophyllase; DVR, divinyl reductase; CHLG, chlorophyll/bacteria chlorophyll a synthase; CPS, ent-copalyl diphosphate synthetase; KS, ent-kaurene synthase; GA20OX, gibberellin 20-oxidase; GA3OX, gibberellin 3-oxidase.