Highlights
-
•
Some common epigenetic regulations exist between coronary artery disease (CAD) and non-small cell lung cancer (NSCLC).
-
•
VEGFA and AIMP1 both are up-regulated/ down-regulated in a similar pattern in both CAD and NSCLC.
-
•
Several DNA damage-repair factors (e.g., BRCA1, ERCC1, XPF, RAD51 etc.) and R-loops are involved in CAD and NSCLC.
Keywords: Epigenetics, Histone methylation, VEGFA, AIMP1, BRCA1, R-loop
Abstract
The effect of epigenetics in coronary artery disease and Non-small cell lung cancer (NSCLC) is presently developing as a significant vital participant at various levels from pathophysiology to therapeutics. We would like to find out the conjunction of some regular epigenetic regulations which decides the example of either acetylation/deacetylation or methylation/demethylation on various gene promoters associated with their pathogenesis. Expressions of some of the genes (e.g., VEGFA, AIMP1, etc.) are either up regulated or down regulated in a similar pattern where several DNA damage (e.g. H2A.X) and repair factors (e.g. BRCA1, RAD51, ERCC1, XPF), Transcription coupled DNA repair factor, Replication proteins are involved. Additionally, epigenetic changes, for example, histone methylation was found unusual in BRCA1 complex in CAD and in the NSCLC patients. Epigenetic therapies such as CRISPR/Cas9 mediated knockout/overexpression of specific gene (BRCA1) showed promising changes in diseased conditions, whereas it affected the R-loop formation which is vulnerable to DNA damage. Involvement of the common epigenetic mechanisms, their interactions and alterations observed in our study will contribute significantly in understanding the development of novel epigenetic therapies soon.
Introduction
Cardiovascular diseases contain plenty of diverse pathological conditions, among which a remarkable feature relates to coronary artery disease (CAD) and its significant hazardous inconvenience [1]. Lung cancer is also a serious health issue and cause of mortality preoccupying worldwide prevailing in men as well as women [2]. The most prevalent type is non-small cell lung cancer (NSCLC) consisting of 85% [3]. Coronary artery disease and non-small cell lung cancer often coexists and cause substantial public health and economic burden worldwide. There is an intrinsic interplay between NSCLC and CAD, in the form of shared etiology and pathophysiological mechanisms. It considered diseases that emerge from different variables in their development and shared several risk factors such as inflammation, oxidative stress, uncontrolled proliferation [4], may be connected including; nutritional, psychosocial, and environmental conditions to start the change of cells in the coronary artery wall and the cycle of atherogenesis, stresses numerous similitudes with the movement of the neoplastic process towards cancer. It has been hypothesized that lung cancer is associated with increased cardiovascular risk, especially the risk for coronary heart disease and stroke [5], [6]. Patients with suspected or known CAD often have a higher lung cancer risk as well [7]. Among patients going through coronary revascularization, more prominent than half of the passings are expected for non-cardiac causes, of which 20% are identified with neoplasia [8]. CD4+ T helper cells contribute to regulate immune responses to tumors, cardiovascular illness, and are significant in organizing overall immune responses. Thus revealing their mechanism in several life-threatening diseases might lead us to understand the cause of the diseases and development of therapies.
The nonstop inability to fix DNA damage can bring about the collection of lethal DNA damage, prompting prolonged p53 activation and p53-mediated activity, leading to cardiomyocyte apoptosis, and resulting in cardiac dysfunction [9]. Along with this, dysregulation of R-loop digestion hinders genomic stability, replicative senescence, and epigenetic balance. Alternately, unresolved R-loops cause damage to the genome by ultimately disrupting transcription which invokes activation of transcription-coupled nucleotide excision repair factors that cleave R-loops through the induction of DNA single strand breaks and double strand breaks [10].
The effect of epigenetics in CAD and NSCLC is presently developing as a significant vital participant at various levels from pathophysiology to therapeutics, suggesting the possibility of complex interplay between genes through epigenetic modifications which may account for their frequent coexistence. The way that epigenetic deviations, in contrast to hereditary transformations, are possibly reversible and can be re-established to their ordinary state by epigenetic treatment make such activities favourable and restoratively significant.
However, there has been limited success in correlating NSCLC with CAD in terms of epigenetic changes and epigenetic commitment in clinical practice or therapeutics up to now. Therefore, to support a hypothesis of an intrinsic interplay between NSCLC and CAD, we evaluated some common epigenetic regulators of NSCLC and CAD which act on different genes leading to various molecular mechanisms, to better understand the association of these two disorders.
Materials and methods
Cells
With the approval of our Institutional Ethics Committee of SRM Medical college Hospital, obtained written informed consent, we prospectively included patients of each diseased condition (CAD and NSCLC) from two centres aged from 35 to 65 years from July 2018 to January 2020. Patients underwent coronary computed tomography angiography (CCTA) for detection of coronary artery disease and chest X-ray followed by pathology proven for NSCLC. Age-matched non-smokers were taken for all the groups (control subjects, CAD and NSCLC patients) in this study. All blood samples were handled as per protocol accredited by the Institutional Ethics Committee. Age, gender, pathological and medication details were recorded for analysis in molecular studies involved in this project. Peripheral blood mononuclear cells (PBMCs) isolation was done by density gradient centrifugation using Ficoll-paque (GE healthcare); followed by CD4+ T cell isolation using CD4+ isolation Kit human (Miltenyi Biotec) through Magnetic-activated cell sorting [11]. As we are studying the epigenetic regulations associated with the diseases our work is mainly concentrated on CD4+ T helper cells isolated from blood. We have not studied tissue biopsies and tissue infiltrating T cells.
Quantitative real-time PCR
This assay was performed on total RNA isolated from CD4+ T helper cells of control subjects, CAD and NSCLC patients as described. The primers for mRNAs utilized in this study are shown in supplementary Table 1 and GAPDH was used for normalization. Derived Ct values were used to calculate fold changes of gene expression as previously described [10].
RT2 profiler PCR array
Quick-RNATM Miniprep (Zymo Research) kit was used to isolate mature RNA as per manufacturer's protocol and we used spectrophotometer to determine its quality as pure RNA. It is then converted into cDNA by the reverse transcription process using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) followed by RT² Profiler PCR Array (QIAGEN). RT² SYBR® Green qPCR Mastermix (QIAGEN) has been used for this experiment according to manufacturer's instruction. Samples were marked properly as control group, test group 1 as CAD and test group 2 as Cancer. CT values were produced and normalization of CT values were done based on a/an automatic selection from HKG panel of reference genes and used delta delta CT method to calculate fold change. Delta delta CT method is the calculation between gene of interest (GOI) and an average of reference genes (HKG), followed by delta-delta CT calculations (delta CT (Test Group)-delta CT (Control Group)). Scatter plots and heat map were generated after calculating fold change using 2^ (-delta delta CT) formula which was acquired from the QIAGEN web portal at GeneGlobe.
Gene-gene interaction and network analysis
A list of up and down regulated genes from CAD (Group 1) and NSCLC (Group 2) was analysed further using a bioinformatics approach. To understand the impact of dysregulation on the disease pathogenesis, gene-gene association was evaluated first using GeneMANIA server for both the disease groups [12]. The interaction table for both the groups were then downloaded and formatted, followed by cluster analysis using MCODE [13] in the Cytoscape software. To test the functional attributes of the differential gene expression patterns, we integrated Gene Ontology (GO) terms with metabolic pathway information using the Cytoscape plug-in ClueGO [14]. The procedure has been detailed in Supplementary Fig. 1.
Chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR)
Chromatin was isolated from fixed peripheral blood mononuclear cells from CAD, NSCLC patients and control subjects followed by digestion with micrococcal nuclease (MNase). It was then immunoprecipitated using specific antibodies/ reagents. After that it was incubated overnight and washed using RIPA buffer. DNA was isolated by phenol-chloroform-isoamylalcohol and stored at -20°C as per µChIP assay protocol. These samples were then used for q-PCR on the target genes to acquire Ct values where control IgG-ChIP nonspecific signals were subtracted from which percent enrichment was calculated using specific formula and represented on a graph [11].
Co-Immunoprecipitation (co-IP) and immunoblotting (IB)
These assays were executed using the commercial reagents (supplemental data) as mentioned. [11]
Histone methylation assay
This assay was performed to test the in vitro H3 lysine methyltransferase activity of immunoprecipitated (IP) BRCA1 as described [11]. Hela core histones and anti-RbBP5 Ab served as the positive control, whereas IgG-IP served as the negative control.
CRISPR/Cas9-mediated knock-out (KO) and Overexpression (OE) assay
CRISPR/Cas9 mediated knockout was done to suppress the expression of endogenous BRCA1 in primary human CD4+T helper cells isolated from control subjects, NSCLC and CAD patients. Transfection was performed using CRISPR/Cas9 knock-out (KO) plasmids (Santa Cruz) and their control plasmid (Santa Cruz) utilizing all the supplied reagents as per the manufacturer's protocol. Transfection was done successfully which was verified by flow cytometry and transfection efficiency was checked by GFP staining. BRCA1 primary antibody (Santa Cruz) and secondary antibody, Goat anti-Rabbit IgG (H+L) APC (Santa Cruz) was used for this purpose. Human tagged ORF clone of BRCA1 gene (Origene) was used to overexpress the BRCA1 gene using AmaxaTM Cell Line NucleofectorTM Kit V (Lonza) and Amaxa nucleofector 2b. After that flow cytometry was done using BRCA1 primary antibody (Santa Cruz) and a secondary antibody, Goat anti-Rabbit IgG (H+L) Alexa Fluor 594 (Invitrogen) to verify successful overexpression of the particular gene [15].
DNA-RNA immunoprecipitation and RT-PCR
DNA-RNA immunoprecipitation (DRIP) was performed as mentioned [16].
Statistical analysis
All results represent the average of 3–5 separate in vitro experiments. In all assay a value represents the mean of three individual observations and presented as standard error mean (SEM). Statistical significance was established by Tukey: Compared all pairs of columns test based one-way ANOVA using INSTAT3 Software.
Results
Expression of several DNA damage repair pathway genes involved in CAD and NSCLC
Mis-repaired DSBs can result in loss of genetic information, potentially harmful mutations or chromosomal rearrangements, which leads to cancer development [17]. DNA damage repair factor BRCA1, a caretaker of genomic stability and also responsible for cardiomyocyte survival [18], associated with RAD51 and DSBs repair factors ATM, ATR are found to be (Fig. 1A, 1B) expressed less in CAD and NSCLC patients Th cells when compared to control subjects as they are the key players in repairing DNA damage, whereas, the expression of DNA damage factor, H2AFX and tumor suppressor gene P53 is more in the person suffering from NSCLC and CAD when tallied to control subjects. Deregulation of DDR pathways can also contribute to the development of genomic instability, which can accelerate the epigenetic alterations to drive tumor development as well as CAD.
Fig. 1.
mRNA expression profile of the DNA repair and damage factors. (A,B): RT-qPCR mRNA expression profile of the indicated candidate genes (BRCA1, RAD51, ATM, ATR, H2AFX, P53) in control subjects, CAD and NSCLC patients’ CD4 + T helper cell. Data were generated from 3 biological replicates with bars indicating SEM. ***p <0.001, one-way ANOVA statistical analysis that compares values in control subjects with those in CAD and NSCLC patients.
RT2 Profiler PCR Array revealed common up-regulated and down-regulated genes involved in CAD and NSCLC patients CD4+ T cells
Since both diseases are characterized by a chronic inflammatory process [4], our RT2 Profiler PCR array reveals the expression of several inflammatory genes which are commonly involved in both CAD and NSCLC. The abnormal fold regulation of all the genes involved were listed (Table 1).
Table 1.
RT2 Profiler PCR Array analysis report showing gene with fold regulation between coronary artery disease (CAD) and non-small cell lung cancer (NSCLC) CD4+ T helper cells compared to control.
| Genes | CAD (compared to control) | NSCLC (compared to control) |
|---|---|---|
| AIMP1/EMAP2 | ↓↓ | ↓↓ |
| BMP2 | ↑↑ | ↑↑ |
| C5 | ↑ | ↑ |
| CCL1 | ↑ | ↑ |
| CCL11 | ↑ | ↑ |
| CCL13 | No change | ↑ |
| CCL15 | ↑ | ↑ |
| CCL16 | ↑ | ↑ |
| CCL17 | ↑ | ↑ |
| CCL2 | ↑ | ↑ |
| CCL20 | ↑ | ↑ |
| CCL22 | ↑ | ↑ |
| CCL23 | ↑ | ↑ |
| CCL24 | ↑ | ↑↑ |
| CCL26 | ↑ | ↑ |
| CCL3 | ↑ | ↑ |
| CCL4 | ↑ | ↑ |
| CCL5 | ↑ | ↑ |
| CCL7 | ↑ | ↑ |
| CCL8 | ↑ | ↑ |
| CCR1 | ↑ | ↑ |
| CCR2 | ↑ | ↑ |
| CCR3 | ↑ | ↑ |
| CCR4 | ↑ | ↑ |
| CCR5 | ↑ | ↑ |
| CCR6 | ↑ | ↑ |
| CCR8 | ↑ | ↑ |
| CD40LG | ↑ | ↓↓ |
| CSF1 | ↑↑ | ↑↑ |
| CSF2 | ↑ | ↑ |
| CSF3 | ↑ | ↑ |
| CX3CL1 | ↑ | ↑ |
| CX3CR1 | ↑↑ | ↑↑ |
| CXCL1 | ↑↑ | ↑ |
| CXCL10 | ↑ | ↑↑ |
| CXCL11 | No change | ↑ |
| CXCL12 | No change | ↑ |
| CXCL13 | ↑ | ↑ |
| CXCL2 | ↑↑ | ↑ |
| CXCL3 | ↑ | ↑ |
| CXCL5 | ↑ | ↑ |
| CXCL6 | ↑ | ↑ |
| CXCL9 | No change | No change |
| CXCR1 | ↑↑ | ↑ |
| CXCR2 | ↑↑ | ↑ |
| FASLG | No change | ↓↓ |
| IFNA2 | No change | ↓↓ |
| IFNG | No change | ↓↓ |
| IL10RA | ↑↑ | ↑ |
| IL10RB | ↑↑ | ↑ |
| IL13 | ↑ | ↑ |
| IL15 | ↑ | No change |
| IL16 | ↑↑ | No change |
| IL17A | ↑ | ↑↑ |
| IL17C | ↑ | ↑↑ |
| IL17F | No change | ↑ |
| IL1A | ↑ | ↑↑ |
| IL1B | ↑↑ | ↑ |
| IL1R1 | ↑ | ↑ |
| IL1RN | ↑↑ | ↑ |
| IL21 | ↑ | ↑↑ |
| IL27 | ↑ | ↑↑ |
| IL3 | ↑ | ↑↑ |
| IL33 | ↑ | ↑↑ |
| IL5 | ↑ | ↑↑ |
| IL5RA | ↑ | ↑↑ |
| IL7 | No change | ↓↓ |
| CXCL8 | ↑↑ | ↑↑ |
| IL9 | ↓ | ↓ |
| IL9R | ↓ | ↓ |
| LTA | ↓ | ↓ |
| LTB | ↓↓ | ↓↓ |
| MIF | ↑ | ↑ |
| NAMPT | ↑ | ↑↑ |
| OSM | ↑↑ | ↑↑ |
| SPP1 | No change | ↓↓ |
| TNF | No change | ↓↓ |
| TNFRSF11B | No change | ↓ |
| TNFSF10 | ↑↑ | ↓ |
| TNFSF11 | ↑ | ↓ |
| TNFSF13 | No change | ↓↓ |
| TNFSF13B | ↑ | ↓↓ |
| TNFSF4 | ↑ | ↓ |
| VEGFA | ↑↑ | ↑↑ |
| ACTB | No change | No change |
| B2M | ↑ | No change |
| GAPDH | ↑ | No change |
| HPRT1 | ↓ | No change |
| RPLP0 | ↑ | No change |
Scatter plots (Fig. 2A, 2C) and heat maps (Fig. 2B, 2D) showed that expressions of some of the genes are either up regulated/down regulated in a similar pattern in both CAD and NSCLC patients Th cells. Among these, VEGFA, AIMP1, BMP2, CSF1, OSM and several interleukins found to be commonly dysregulated.
Fig. 2.
Identification of common upregulated/downregulated genes. (A): The normalized expression of genes between control group and test group 1 (CAD group) was plotted in a scatter plot to compare and visualize gene expression changes of every gene on the array where unchanged gene expression was indicated by the central line and fold regulation threshold by the dotted lines. There are several data points beyond the dotted lines which meet the selected fold regulation threshold in the upper left and lower right sections. (B): Gene expression changes between the control group and test group 1 (CAD group) is represented by a heat map where fold-change values greater than one indicates a positive- or an up-regulation and fold-change values less than one indicate a negative or down-regulation. (C): The normalized expression of genes between control group and test group 2 (Lung CA group) was plotted in a scatter plot to compare and visualize gene expression changes of every gene on the array where unchanged gene expression was indicated by the central line and fold regulation threshold by the dotted lines. There are several data points beyond the dotted lines which meet the selected fold regulation threshold in the upper left and lower right sections. (D): Gene expression changes between the control group and test group 2 (Lung CA group) is represented by a heat map where fold-change values greater than one indicates a positive- or an up-regulation and fold-change values less than one indicate a negative or down-regulation.
Vascular endothelial growth factor A (VEGFA), is the lead player of angiogenesis in cancer and also in hypoxia [19]. According to our findings VEGFA was highly expressed in both CAD and NSCLC patients Th cells as compared with the control subjects. AIMP1 prompts both endothelial cell death and immunity-promoting cytokine production, it is relied upon to control tumor development through a dual mechanism. Consistent with this, AIMP1 was found to be downregulated in CAD and NSCLC patients Th cells than the control subjects.
Correlation between CAD and NSCLC diseased state
At a p-value of 0.05, 80 genes from CAD and 85 genes from NSCLC were observed to be differentially regulated. Based on fold change, the majority of these genes were up-regulated, amongst which 59 were common in both the disease groups. In CAD, 384 and NSCLC a total of 468 interactions were observed with a high overall interaction score of >0.9. Cluster analysis for the dysregulated genes in CAD and NSCLC showed the highest clustered network (score=40.02 and 41.54) had 46, 49 genes with over 900, 997 interaction amongst them (Supplementary Figs. 2, 3). Separately, the downregulated genes showed similar level of interaction too (Supplementary Figs. 4, 5).
Inflammatory pathway association
There is a total of 213 genes that were reported to be involved in the positive regulation of inflammatory response. Association studies with the NSCLC up-regulated genes generated a highly clustered network of 82 nodes, each with an average of 36 interactions. With the CAD disease state, the total nodes in the entire network were 90. Compared to NSCLC, CAD displayed a higher average number of interactions (38) per node. 838 interacting nodes were seen to be co-expressed between CAD and inflammatory pathway genes compared to 690 in NSCLC. Inflammatory pathway was involved with the largest number of upregulated genes. The combined cluster of inflammatory pathway and up-regulated genes in NSCLC and CAD revealed VEGFA to be forming an integral part of the merged cluster. This consensus network was generated from MCODE analysis. Similarly, AIMP1 was a common node for co-expressed gene network of inflammatory pathway, including NSCLC and CAD down-regulated genes (Fig. 3A, 3B), hence we have focused our study on these particular two genes i.e. VEGFA and AIMP1. Crucial immunomodulators such as IFNG, IFNA2 responsible for anti-proliferative and induction of innate immune system were found to be downregulated. The underlying connection remains unclear in spite of a wide range of common cellular processes reported here.
Fig. 3.
(A) Merged co-expressed gene network of inflammatory pathway including NSCLC and CAD up-regulated genes showing VEGFA gene as a common node. (B) Merged co-expressed gene network of inflammatory pathway including NSCLC and CAD down-regulated genes showing AIMP1 gene as a common node.
Aberrant expressions of DNA damage and repair factors at VEGFA and AIMP1 loci
Once a DNA damage event is detected, it can be repaired by one or several of the DNA repair pathways and involves several DNA damage (e.g. H2A.X), and repair factors (e.g. BRCA1, RAD51, ERCC1, XPF), Transcription coupled DNA repair factors (e.g. CSA, CSB) and Replication proteins (e.g. TOP1, RPA70). ChIP-qPCR data showed the percent enrichment of the proteins in AIMP1 and VEGFA gene loci where DNA repair factors, BRCA1 and RAD51 are recruited to DNA more at VEGFA and AIMP1 loci, however, in contrast, DNA damage factor, gH2A.X is recruited less (Fig. 4A, 4B) in control subjects Th cells than the CAD and NSCLC patients Th cells. ERCC1, XPF, CSA, CSB, TOP1 and RPA70 are recruited to DNA more at VEGFA and AIMP1 loci (Fig. 4C, 4D) in control subjects Th cells than the CAD and NSCLC patients Th cells. BRCA1 was discovered to be related with enormous protein complexes that contain DSB fix and mismatch-repair enzymes as well as ATM [20]. Reports indicated that during DNA damage BRCA1 co-localizes with phosphorylated H2AFX [21]. Decreased expression of CSB has already been demonstrated to increase the risk of lung cancer [22]. Through Co-Immunoprecipitation the protein-protein interaction of the different proteins involved were seen. Hence it can be inferred that the amount of protein interaction in CAD and NSCLC is relatively more similar. The involvement of BRCA1 was found to be comparatively higher in controls than CAD and NSCLC, whereas gH2A.X showed less involvement (Fig. 4E) in control as compared to CAD and NSCLC. RAD51 showed a moderate expression in all, slightly varying in CAD and NSCLC. As DNA repair pathways are very crucial to prevent the accumulation of DNA lesions, abnormal expressions of these factors may promote CAD along with NSCLC.
Fig. 4.
Expression of several DNA damage and repair factors on the identified gene locus. (A, B): ChIP-qPCR was performed on primary Th cells from normal donor and CAD and NSCLC patients with indicated antibodies (BRCA1, RAD51, gH2A.x) at the indicated gene loci (VEGFA and AIMP1). (C,D): ChIP assays were performed on primary Th cells from normal donor and CAD and NSCLC patients with indicated antibodies (CSA, CSB, XPF, TOP1, ERCC1 and RPA70) at the indicated gene loci (VEGFA and AIMP1). Data were generated from 3 biological replicates with bars indicating SEM. ***p <0.001, one-way ANOVA statistical analysis that compares values in control subjects with those in CAD and NSCLC patients. (E): Co-IP was done using antibodies to BRCA1, RAD51 and gH2A.X and immunoblotted (IB) with the Rad51 and gH2A.X antibody performed on the nuclear extracts isolated from the primary Th cells of control subjects, CAD patients and NSCLC patients.
Histone H3K4 trimethylation is not catalyzed by BRCA1 in CAD and NSCLC patients Th cells
In the quickly advancing field of cancer epigenetics, histone modifications and alterations in histone methylation patterns have been indicated as an important part of the epigenetic regulations in cancer [23] and also in cardiovascular diseases. Our HMTase assay revealed that BRCA1 complex catalyzes histone H3K4 trimethylation in control subjects Th cells (Fig. 5) but not in CAD patients Th cells like NSCLC patients Th cells. According to several studies, the level of methylation of histone 3lysine 4 (H3K4me3) was elevated in lung cancers and played a role in aberrant transcriptional regulation. Furthermore, their ChIP-seq research revealed that H3K4me3 levels near transcriptional start sites in lung cancer cell lines were elevated. In individuals with CHD, mutations in multiple genes that generate and read methylation H3K4 have been discovered. [24], [25], [26]
Fig. 5.
Analysis of histone H3K4 trimethylation activity by histone H3 HMTase assay: Histone H3 methyltransferase activity of BRCA1 IP with anti-BRCA1 Ab from the nuclear fractions of CD4+Th cells isolated from CAD and NSCLC patients. Material IP with anti-RBBP5 Ab served as positive control. Immunoblotting was done with the indicated series of Abs. The data represents at least five independent assays from five separate CAD and NSCLC patients and five separate control subjects.
CRISPR-Cas9 knockout and Overexpression of BRCA1 gene and its expression on VEGFA and AIMP1 loci
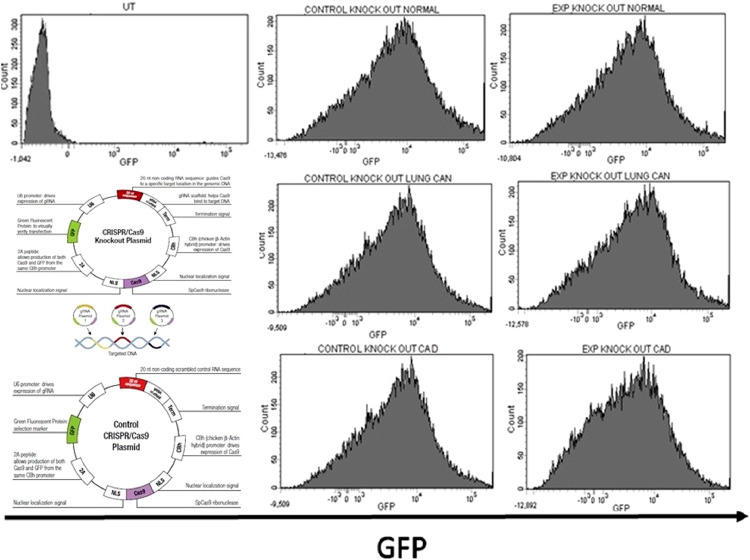
The histogram plots (Fig. 6A), showing successful knock-out of BRCA1 gene and Fig. 6B shows the overexpression of BRCA1 in CD4+ T helper cells isolated from CAD, NSCLC as well as control.
Fig. 6.
CRISPR-Cas9 mediated knockout of abnormally expressed BRCA1 gene: Flow cytometry. (A) Histograms show the transfection efficiency of the indicated CRISPR construct in CD4+T helper cells isolated from control subjects, CAD and NSCLC patients. (B) Histograms show BRCA1 expression in BRCA1 knock-out (KO) and control KO CD4+T helper cells isolated from control subjects, CAD and NSCLC patients. (C): Flow cytometry. Histograms show over-expression of BRCA1 in CD4+T helper cells isolated from NSCLC and CAD patients.
We observed a depletion of BRCA1 leads to the suppression of expression of BRCA1 gene in VEGFA and AIMP1 loci (Fig. 7A, 7B), and are recruited to DNA more after overexpression of BRCA1 as evidenced from our µChIP assays. After successful depletion of BRCA1, it has shown less enrichment in VEGFA and AIMP1 gene loci which confirm that BRCAness not only in NSCLC but also in coronary artery disease. Studies have shown that the recently developing gene editing technology CRISPR/Cas9 has held a number of points of interest over different procedures, including its straightforward plan, simple activity, great explicitness, and high proficiency. Therefore, repairing/silencing the expression of tumor genomes or specific proteins is important for [27] tumor therapy, modifications [28,29], exploring the mechanisms underlying tumor occurrence, development, and metastasis.
Fig. 7.
Analyzing expression of P53 gene after knockout. (A, B): ChIP-qPCR was performed on primary Th cells from control subjects and CAD and NSCLC patients with BRCA1 antibody at the indicated gene loci (VEGFA and AIMP1). (C, D): P53 gene expression changes through RT-qPCR based mRNA expression in NSCLC and CAD patients’ CD4+ T helper cells in comparison to control subjects. Data were generated from 3 biological replicates with bars indicating SEM. ***p <0.001, **p <0.01, *p <0.05, one-way ANOVA statistical analysis that compares values in control subjects with those in CAD and NSCLC patients.
Increased fold change of P53 gene after overexpression of BRCA1 and decreased fold change was observed post suppression of endogenous BRCA1
TP53 has multiple faucets to its role in the maintenance of genomic stability, including responding directly to DNA damage to promote repair and cell cycle arrest, the transcriptional regulation of DNA repair genes and the induction of apoptosis. In the absence of TP53 cells accumulate DNA damage and resist cell death. TP53 has been shown to be mutated in around 50% of all NSCLCs [28]. Our mRNA expression profile of P53 gene represents the fold change where we can observe that post depletion of BRCA1, it is expressed less in CAD (Fig. 7C) and in NSCLC (Fig. 7D) patients when compared to controls, whereas BRCA1 overexpression causes increased fold change for P53 in CAD and NSCLC patients, which shows the link between BRCA1 and P53 that work as a DNA damage repair cascade.
P53 is mainly associated with tumor suppressor activity. Because BRCA1 is also a gatekeeper of cardiac function and survival, one study found that cardiomyocyte BRCA1 deletion causes defective DNA double-strand break repair and activated p53-mediated proapoptotic signalling, resulting in increased cardiomyocyte apoptosis. Deletion of the p53 gene, on the other hand, recovers BRCA1-deficient mice from heart failure, demonstrating yet again the relationship between these two genes. [18]
BRCA1 depletion provokes R-loop accumulation in Human CD4+ Th cells
Emerging evidence suggests that the crosstalk exists between R-loops and epigenetic modifications of chromatin. Certain chromatin modification features limit R-loop development on the one hand, while their establishment can increase chromatin dynamics by modifying epigenetic changes on the other [30].
In DRIP-qPCR, we showed that R-loops formed at a high level on the VEGFA and AIMP1 locus after successful depletion of endogenous BRCA1, whereas the R-loop percent enrichment is at low level after overexpression of BRCA1 (Fig. 8A, 8B).
Fig. 8.
Expression of R-loops through DRIP-qPCR: (A, B): R-loop prevalence on VEGFA and AIMP1 gene loci in control subjects, NSCLC and CAD patients’ CD4 + T helper cells. Data were generated from 3 biological replicates with bars indicating SEM. ***p <0.001, *p <0.05, one-way ANOVA statistical analysis that compares values in control subjects with those in CAD and NSCLC patients.
As R-loops are co-transcriptional items, these discoveries, recommend that in diseased condition nascent transcription is occurring at this BRCA1 gene locus to fuel R-loop production. Since unrepaired R-loops cause DSBs, we explored the impact of increased R-loops on genomic stability in knockout condition. We conclude that BRCA1 deficiency or abnormality results in DNA damage through accumulation of RNA/DNA hybrids, but when introduced through overexpression, R-loop formation has been decreased.
R-loop-induced epigenetic changes are likely context-dependent and determined by complex regulatory mechanisms, such as where they form (e.g. promoters, enhancers, or termination regions), what causes them to form (e.g. mRNP biogenesis, DNA topoisomerase, trinucleotide-repeat, or antisense transcription), and what characteristics they have (e.g. length, additional structures of single-stranded DNA within R-loop and so on) [30].
Discussion
The results showed a significant correlation among CAD and NSCLC, suggesting the possibility of shared pathophysiology of the diseases. NSCLC and CAD are both associated with the hallmarks of inflammation. All immune cells in our body play important role in each aspects. For inflammatory diseases, T cells are the key cell types involved among which CD4+ T cells primarily mediate anti-tumor immunity by providing help for other cell types. The CD4+T cells carry out multiple functions, ranging from activation of cells of the innate immune system, B-lymphocytes, cytotoxic T cells, macrophages, monocytes as well as nonimmune cells via secretion of effector cytokines such as interferon-γ (IFNγ) and tumor necrosis factor-α (TNFα) [31], and also play critical role in the suppression of immune reaction. Other immune cells such as dendritic cell and NK cells help in priming T cell responses and also provide signals for the differentiation of CD4+ T cells into effector T cell populations, improve maturation and cross-presentation of DCs and are thereby able to promote T cell responses. PMN cells work as accessory cells for T-cell activation and thus making CD4+T cells as the central player of inflammatory response, so we have taken particularly CD4+T cell type (excluding other cell types) in this study. We have identified the common up-regulated/down-regulated genes and the correlation between NSCLC and CAD, was most significant in the CD4+ T helper cells gene expression profiles. This work is highly impacted to study the CD4+ T cell mediated inflammatory regulation as we have tested mainly CD4+ T helper cells but not the other subsets of T helper cells. We have taken whole CD4+ T helper cell population which is associated with diseased state. Previous studies have reported cases of CAD progressing to various cancer pathogenesis [32]. To explore a possible disease progression to NSCLC, understanding both up and down-regulated gene sets become crucial. Initially, it was observed that all the up-regulated CAD disease state genes were identical to those found in NSCLC. The present study showed that up-regulated genes in CAD and inflammatory genes presented a high degree of cluster in the gene-gene interaction. Interestingly, we were able to observe up-regulated genes from NSCLC to also contribute to this network. Analysis of GO terms later revealed a positive correlation of these genes mainly involved in pathways of angiogenesis, inflammatory response and receptor signalling, cytokine mediated signalling and aging amongst other processes suggesting their important role in the pathogenesis of CAD and NSCLC [4]. Inflammatory response genes PI3K, AKT, IGF1R amongst others were found to be linked with NSCLC up-regulated genes [33]. In the downregulated state, both disease state genes were highly connected with the inflammatory pathway genes. Since VEGF is the key arbiter of angiogenesis, anomalies of its expression can possibly be significant in disease progression. Moreover, biopsies speaking to an enormous human tumor types have been appeared to show increased expression of VEGF, and has been perceived to be crucial to tumorigenesis and disease development in a wide scope of human diseases [19] and its overexpression is related with poor prognosis and demise from metastasis [34], [35], [36]. AIMPs contribute in several biological processes [37], [38], [39], [40] and could distinctively cooperate to ten different cancers, while there has been advancement in understanding the function of AIMP1 in cancer [37,41,42]. AIMP1 is also found to be abundant in tissues going through apoptosis in the mouse embryo and atherosclerotic injuries of the human aorta [34]. Toward this end, we compared the gene expression profiles of NSCLC and CAD to show the association in-between them, and tried to explain the shared pathophysiology. To find out the possible molecular mechanisms behind this dysregulation, we have analysed several DDR genes involved. DSBs are found to be significantly higher in patients with CAD [43] and deficiencies in DDR mechanisms have been shown to be contributing factors in many stages of tumor development. The correct functioning of DDR plays an important role in many aspects and is linked to the induction of replication stress and DNA damage, caused by abnormal replication due to aberrant oncogene activation [44,45]. Hence the involvement of aberrant expressions of several DNA damage/repair factors has been identified on AIMP1 and VEGFA gene locus provides clear evidence of the abnormalities involved which clarifies the similarities in both the diseases and revealed the mechanism lying behind it involving several molecular pathways. Proof suggests that the disruption of tumor suppressor genes, for example, p53 is related to expanded VEGF expression [46]. So, again our mRNA expression data of several repair factors involved proved their loss of normal function in both the diseases along with the abnormality in histone modification. Histone methylation is an emerging epigenetic mechanism for regulating gene transcription. Interplay among cardiac transcription factors and histone lysine modifiers plays important role in heart development. Aberrant expression and mutation of the histone lysine modifiers during development and in adult life influence the response of adult hearts to pathological stresses. Dysregulation of histone methylation are closely linked to clinical outcomes in lung cancer patients through a variety of cellular pathways relating proliferation, invasion, (Epithelial-mesenchymal transition) EMT etc. [24]. Although the loss of the normal DNA repair machinery and accumulation of epigenetic aberration can drive the tumor progression, it also provides a targetable defect for therapeutic intervention. Regulators of the DDR have therefore become attractive targets for cancer therapy. Therefore, it is significant to rectify and edit the genome or by introducing overexpression using CRISPR/Cas9 technology for tumor epigenetic disorders at gene level approaches for treating BRCA-proficient malignancies like NSCLC [47]. Several proteins involved in the DNA damage response have been identified as emerging targets for cancer treatment, including; PARP1, ATR, and Chk1. While PARP inhibitors use in BRCA1/2 mutated tumors remains the best characterized treatment based on other mutations found in lung cancer, including mKRAS and EGFR mutations [48]. Thus it is obvious why many cancer patients have active heart disease and many heart patients have active cancers. Patterns of epigenetic modification vary among individuals, in different tissues within an individual, and even in different cells within a tissue. So aberrantly functioning CD4+ cells are associated with the development of multiple diseases but it is also important to study other cell types as well which may influence to understand disease epigenetics. Revealing the epigenetic systems along with their communications, and changes utilizing translational approaches vows to make a significant contribution to our comprehension of CADs and NSCLC which may prompt the advancement of novel epigenetic treatments soon.
CRediT authorship contribution statement
Sudeshna Rakshit: Data curation, Formal analysis, Methodology, Software, Writing – original draft. Jithin S. Sunny: Software, Formal analysis. Melvin George: Resources. Luke Elizabeth Hanna: Resources. Koustav Sarkar: Conceptualization, Supervision, Visualization, Resources, Writing – review & editing.
Declaration of Competing Interest
None
Acknowledgement
This research was supported by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Govt. of India (sanction order No. “ECR/2016/000965”).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101189.
Appendix. Supplementary materials
References
- 1.Udali S., Guarini P., Moruzzi S. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol. Asp. Med. 2013;34(4):883–901. doi: 10.1016/j.mam.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen B., Zeng C., Ye Y. Promoter methylation of TCF21 may repress autophagy in the progression of lung cancer. J. Cell Commun. Signal. 2018;12(2):423–432. doi: 10.1007/s12079-017-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zappa C., Mousa S.A. Non-small cell lung cancer: current treatment and future advances. Transl. Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapia-Vieyra J.V., Delgado-Coello B., Mas-Oliva J. Atherosclerosis and cancer; a resemblance with far-reaching implications. Arch. Med. Res. 2017;48(1):12–26. doi: 10.1016/j.arcmed.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Yuan M., Li Q.G. Lung cancer and risk of cardiovascular disease: a meta-analysis of cohort studies. J. Cardiothorac. Vasc Anesth. 2018;32(1):e25–e27. doi: 10.1053/j.jvca.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 6.Cuddy S., Payne S., Murphy D., Dunne R., Groarke J. 52 Prevalence of coronary artery calcification in patients with early stage lung cancer and missed opportunities for cardiovascular risk optimization. Heart. 2017;103:A30. [Google Scholar]
- 7.Gaudio C., Tanzilli A., Mei M., Moretti A., Barillà F., Varveri A., Paravati V., Tanzilli G., Ciccaglioni A., Strano S., Pellegrini M., Barillari P., Pelliccia F. Concomitant screening of coronary artery disease and lung cancer with a new ultrafast-low-dose Computed Tomography protocol: a pilot randomised trial. Sci. Rep. 2019;9(1):13872. doi: 10.1038/s41598-019-50407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieira R.D., Pereira A.C., Lima E.G., Garzillo C.L., Rezende P.C., Favarato D., Hueb A.C., Gersh B.J., Ramires J.A., Hueb W. Cancer-related deaths among different treatment options in chronic coronary artery disease: results of a 6-year follow-up of the MASS II study. Coron. Artery Dis. 2012;23(2):79–84. doi: 10.1097/MCA.0b013e32834f112a. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117(4):305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar K., Han S.S., Wen K.K., Ochs H.D., Dupré L., Seidman M.M., Vyas Y.M. R-loops cause genomic instability in T helper lymphocytes from patients with Wiskott-Aldrich syndrome. J. Allergy Clin. Immunol. 2018;142(1):219–234. doi: 10.1016/j.jaci.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor M.D., Sadhukhan S., Kottangada P., Ramgopal A., Sarkar K., D'Silva S., Selvakumar A., Candotti F., Vyas Y.M. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med. 2010 Jun 23;2(37) doi: 10.1126/scitranslmed.3000813. 37ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar K., Sadhukhan S., Han S.S., Vyas Y.M. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood. 2014;124(23):3409–3419. doi: 10.1182/blood-2014-07-587642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadel J., Athanasiadou R., Lemetre C., Wijetunga N.A., Ó Broin P., Sato H., Zhang Z., Jeddeloh J., Montagna C., Golden A., Seoighe C., Greally J.M. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet. Chromatin. 2015;8:46. doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber A.M., Ryan A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Shukla P.C., Singh K.K., Quan A., Al-Omran M., Teoh H., Lovren F., Cao L., Rovira I.I., Pan Y., Brezden-Masley C., Yanagawa B., Gupta A., Deng C.X., Coles J.G., Leong-Poi H., Stanford W.L., Parker T.G., Schneider M.D., Finkel T., Verma S. BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat. Commun. 2011;2:593. doi: 10.1038/ncomms1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poon R.T., Fan S.T., Wong J. Clinical implications of circulating angiogenic factors in cancer patients. J. Clin. Oncol. 2001;19(4):1207–1225. doi: 10.1200/JCO.2001.19.4.1207. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939. [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida K., Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95(11):866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess J.T., Rose M., Boucher D., Plowman J., Molloy C., Fisher M., O'Leary C., Richard D.J., O'Byrne K.J., Bolderson E. The therapeutic potential of DNA damage repair pathways and genomic stability in lung cancer. Front. Oncol. 2020;10:1256. doi: 10.3389/fonc.2020.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson A.G. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J. Periodontol. 2008;79(8 Suppl):1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q.J., Liu Z.P. Histone methylations in heart development, congenital and adult heart diseases. Epigenomics. 2015;7(2):321–330. doi: 10.2217/epi.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Liu X., Li Y., Quan C., Zheng L., Huang K. Lung cancer therapy targeting histone methylation: opportunities and challenges. Comput. Struct. Biotechnol. J. 2018;16:211–223. doi: 10.1016/j.csbj.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikutake C., Yahara K. Identification of epigenetic biomarkers of lung adenocarcinoma through multiomics data analysis. PloS One. 2016;11 doi: 10.1371/journal.pone.0152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D., Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Shuvalov O., Petukhov A., Daks A., Fedorova O., Ermakov A., Melino G., Barlev N.A. Current genome editing tools in gene therapy: new approaches to treat cancer. Curr. Gene Ther. 2015;15(5):511–529. doi: 10.2174/1566523215666150818110241. [DOI] [PubMed] [Google Scholar]
- 29.Gori J.L., Hsu P.D., Maeder M.L., Shen S., Welstead G.G., Bumcrot D. Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum. Gene. Ther. 2015;26(7):443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 30.Al-Hadid Q., Yang Y. R-loop: an emerging regulator of chromatin dynamics. Acta Biochim. Biophys. Sin. 2016;48(7):623–631. doi: 10.1093/abbs/gmw052. (Shanghai) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay R.E., Richardson E.K., Toh H.C. Revisiting the role of CD4+ T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021;28(1-2):5–17. doi: 10.1038/s41417-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haenszel W., Kurihara M. Studies of Japanese migrants. I. mortality from cancer and other diseases among Japanese in the United States. J. Natl. Cancer Inst. 1968;40(1):43–68. [PubMed] [Google Scholar]
- 33.Molina-Arcas M., Hancock D.C., Sheridan C., Kumar M.S., Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013;3(5):548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goel H.L., Mercurio A.M. VEGF targets the tumour cell. Nat. Rev. Cancer. 2013;13(12):871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berns E.M., Klijn J.G., Look M.P., Grebenchtchikov N., Vossen R., Peters H., Geurts-Moespot A., Portengen H., van Staveren I.L., Meijer-van Gelder M.E., Bakker B., Sweep F.C., Foekens J.A. Combined vascular endothelial growth factor and TP53 status predicts poor response to tamoxifen therapy in estrogen receptor-positive advanced breast cancer. Clin. Cancer Res. 2003;9(4):1253–1258. [PubMed] [Google Scholar]
- 36.Manders P., Beex L.V., Tjan-Heijnen V.C., Geurts-Moespot J., Van Tienoven T.H., Foekens J.A., Sweep C.G. The prognostic value of vascular endothelial growth factor in 574 node-negative breast cancer patients who did not receive adjuvant systemic therapy. Br. J. Cancer. 2002;87(7):772–778. doi: 10.1038/sj.bjc.6600555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko Y.G., Park H., Kim T., Lee J.W., Park S.G., Seol W., Kim J.E., Lee W.H., Kim S.H., Park J.E., Kim S. A cofactor of tRNA synthetase, p43, is secreted to up-regulate proinflammatory genes. J. Biol. Chem. 2001;276(25):23028–23033. doi: 10.1074/jbc.M101544200. [DOI] [PubMed] [Google Scholar]
- 38.Park S.G., Kang Y.S., Ahn Y.H., Lee S.H., Kim K.R., Kim K.W., Koh G.Y., Ko Y.G., Kim S. Dose-dependent biphasic activity of tRNA synthetase-associating factor, p43, in angiogenesis. J. Biol. Chem. 2002;277(47):45243–45248. doi: 10.1074/jbc.M207934200. [DOI] [PubMed] [Google Scholar]
- 39.Park S.G., Shin H., Shin Y.K., Lee Y., Choi E.C., Park B.J., Kim S. The novel cytokine p43 stimulates dermal fibroblast proliferation and wound repair. Am. J. Pathol. 2005;166(2):387–398. doi: 10.1016/S0002-9440(10)62262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S.G., Kang Y.S., Kim J.Y., Lee C.S., Ko Y.G., Lee W.J., Lee K.U., Yeom Y.I., Kim S. Hormonal activity of AIMP1/p43 for glucose homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(40):14913–14918. doi: 10.1073/pnas.0602045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kayton M.L., Libutti SK. Endothelial monocyte activating polypeptide II (EMAP II) enhances the effect of TNF on tumor-associated vasculature. Curr. Opin. Investig. Drugs. 2001;2(1):136–138. [PubMed] [Google Scholar]
- 42.Kim Y.W., Kwon C., Liu J.L., Kim S.H., Kim S. Cancer association study of aminoacyl-tRNA synthetase signaling network in glioblastoma. PLoS One. 2012;7(8):e40960. doi: 10.1371/journal.pone.0040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoudi M., Mercer J., Bennett M. DNA damage and repair in atherosclerosis. Cardiovasc. Res. 2006;71(2):259–268. doi: 10.1016/j.cardiores.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C.L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T.D., Bartek J., Gorgoulis V.G. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 45.Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre' M., Nuciforo P.G., Bensimon A., Maestro R., Pelicci P.G., d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 46.Rak J., Yu J.L., Klement G., Kerbel R.S. Oncogenes and angiogenesis: signaling three-dimensional tumor growth. J. Investig. Dermatol. Symp. Proc. 2000;5(1):24–33. doi: 10.1046/j.1087-0024.2000.00012.x. [DOI] [PubMed] [Google Scholar]
- 47.Abbotts R., Topper M.J., Biondi C., Fontaine D., Goswami R., Stojanovic L., Choi E.Y., McLaughlin L., Kogan A.A., Xia L., Lapidus R., Mahmood J., Baylin S.B., Rassool F.V. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 2019;116(45):22609–22618. doi: 10.1073/pnas.1903765116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung A.W., de Silva T., Bally M.B., Lockwood W.W. Synthetic lethality in lung cancer and translation to clinical therapies. Mol. Cancer. 2016;15(1):61. doi: 10.1186/s12943-016-0546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.