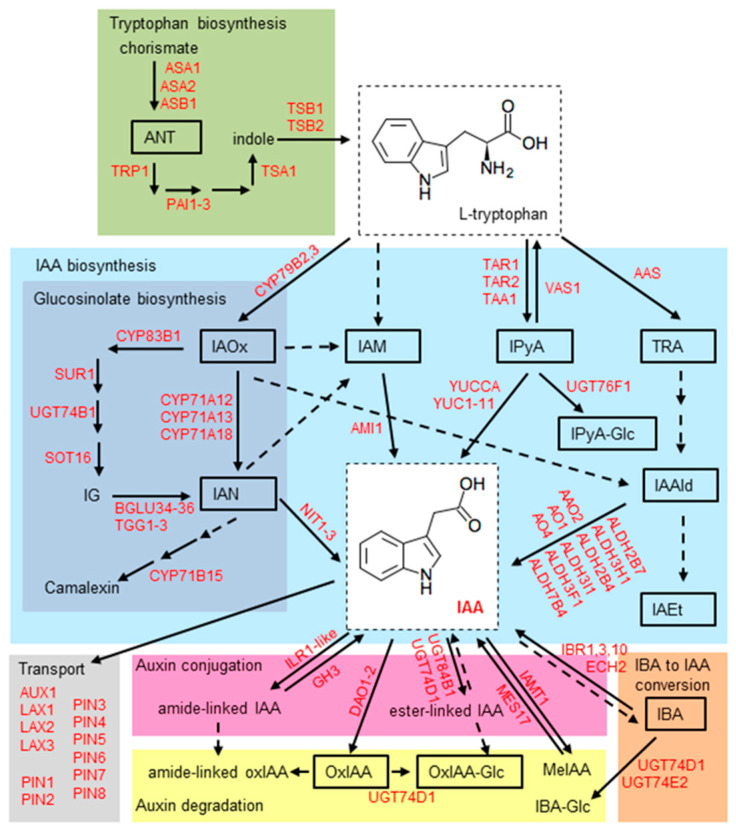
Figure 1.
Scheme of IAA metabolism in Arabidopsis thaliana (Arabidopsis). The pathways involved in auxin metabolism in Arabidopsis are based on the KEGG pathways [13] and data published in [7,14,15,16,17,18,19]. The darker blue panel shows pathways specific for Arabidopsis, Brassica, and Sinapis [20]. Yellow and pink panels indicate pathways for IAA degradation [14] and IAA conjugation [15], respectively. The contribution of the IBA to IAA conversion to the active IAA pool is illustrated by the orange panel [16]. The green panel shows the synthesis of tryptophan from the precursor chorismate via the shikimate pathway [21]. Dashed arrows indicate pathways that may contain multiple enzymatic steps, and where the genes/enzymes are not known. ANT, anthranilate; IAA, indole-3-acetic acid; IAAld, indole-3-acetaldehyde; IAEt, indole-3-acetethanol; IAM, indole-3-acetamide; IAN, indole-3-acetonitrile; IAOx, indole-3- acetaldoxime; IBA, indole-3-butyric acid; IBA-Glc, indole-3-butyryl-1-O-β-d-glucose; IPyA, indole-3-pyruvic acid; MeIAA, indole-3-acetic acid methyl ester; oxIAA, 2-oxoindole-3-acetic acid; oxIAA-Glc, 2-oxoindole-3-acetyl-1-O-β-d-glucose; TRA, tryptamine.