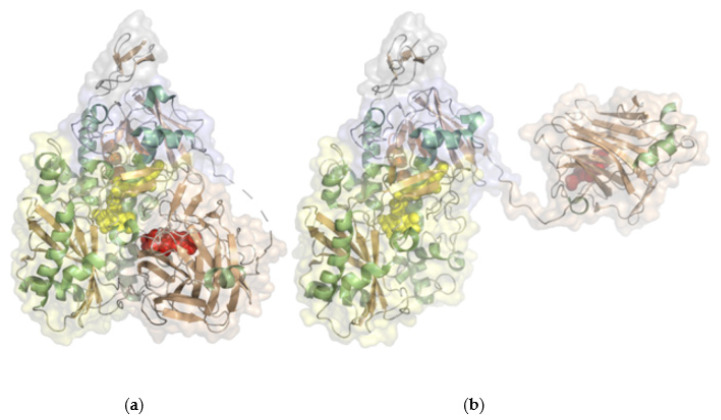
Figure 10.
Crystal structure of (a) Crassicarpon hotsonii cellobiose dehydrogenase in closed conformation (PDB: 4QI6) and (b) Neurospora crassa cellobiose dehydrogenase in open conformation (PDB: 4QI7) with the FAD cofactor (yellow spheres) buried inside the protein shell. The FAD-binding- and substrate-binding subunits are colored light blue and light yellow, respectively. The GMC-oxidoreductase family characteristic dinucleotide binding βαβ-motif (Rossmann-fold) is highlighted in orange. A C-terminal type 1 carbohydrate binding module (CBM_1) is shown “on top” of the structure. The DET-competent cytochrome domain (light red) with the surface exposed heme b cofactor (red spheres) accepts electrons from substrate oxidation from the FAD in the closed conformation and donates them to suitable acceptors in the open conformation.