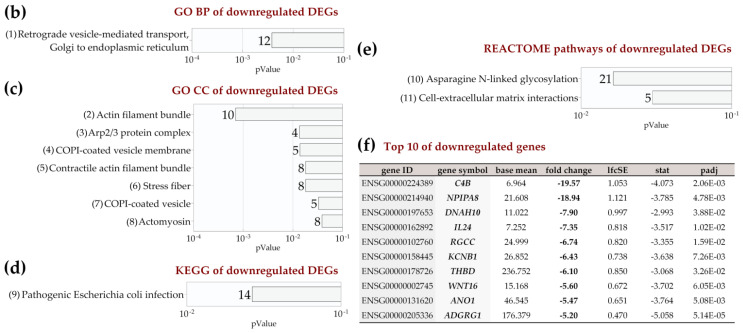
Figure 7.
Functional enrichment analysis of 463 downregulated differentially expressed genes (DEGs) between MSCs preconditioned with Vadadustat and control cells using g:Profiler. The analysis was performed on samples obtained from six populations of hBM-MSCs preconditioned for 6 h with 40 μM Vadadustat compared to MSCs cultured under 20% O2 (control) (a) Results of enrichment analysis presented in the form of a Manhattan plot, where the x-axis shows the functional terms grouped by the color code of source database used, while the y-axis shows the enrichment adjusted p-values in negative decimal logarithm scale. Dots in the graph indicate all enriched terms meeting the significance criterion of p-values < 0.05, while highlighted dots represent terms filtered by the criterion of containing between 5 and 350 genes. The graphs (b–d) show the detailed results of the enriched terms highlighted in the Manhattan plot along with the statistical significance (pValue) and the number of DEGs belonging to the enriched term (placed next to the bar), according to the: (b) gene ontology biological processes (GO BP) (c) gene ontology cellular compartment (GO CC) (d) KEGG (e) REACTOME. (f) Table listing the top ten most highly downregulated genes, ranked by increasing fold change in expression (bold values) including: Ensembl identifier (gene ID), gene symbol, the average of the normalized counts taken over all samples (baseMean), fold change in gene expression (fold change), standard error of the log2FoldChange estimate (lfcSE), Wald statistic (stat) and Benjamini–Hochberg adjusted p-value (padj).