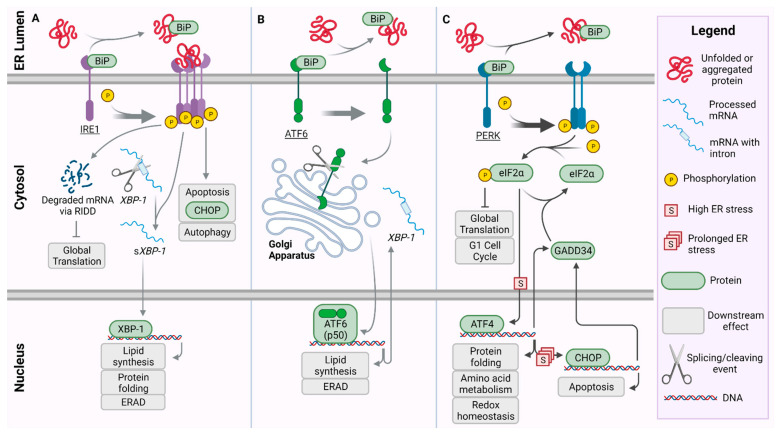
Figure 1.
Pathways of the Unfolded Protein Response (UPR): In the presence of unfolded or aggregated protein, BiP dissociates from the three ER stress sensors: activating inositol requiring enzyme 1 (IRE1) (A), transcription factor 6 (ATF6) (B), and protein kinase RNA (PKR) like ER kinase (PERK) (C). The legend defines graphics for conserved structures between pathways of the UPR. (A) IRE1 forms a phosphorylated tetramer activating a cascade which promotes apoptosis via C/EBP homologous protein (CHOP) and autophagy. Simultaneously, IRE1 promotes the degradation of mRNA to reduce global translation via regulated IRE1-dependent decay (RIDD). The X-box-binding protein 1 (XBP-1) mRNA is additionally spliced, allowing XBP-1 protein to translocate to the nucleus and promote the transcription of UPR target genes for protein folding, lipid synthesis, and endoplasmic-reticulum-associated protein degradation (ERAD). (B) Following release of BiP and activation, ATF6 translocates to the Golgi apparatus where it is cleaved into p50-ATF6. p50-ATF6 serves as a transcription factor for target genes, including those involved in ERAD, lipid synthesis, and XBP-1 which is translocated back to the cytosol. (C) PERK dimerizes and auto-phosphorylates, phosphorylating and inactivating eukaryotic initiation factor 2 alpha subunit (eIF2α). This reduces global translation and causes arrest of the G1 cell cycle via the adaptive UPR. Under high ER-stress conditions, activating transcription factor 4 (ATF4) is upregulated by phospho-eIF2α, then promoting transcription of target genes involved in protein folding, amino acid metabolism, and redox homeostasis. Over prolonged ER-stress, pro-apoptotic CHOP is activated. Accordingly, CHOP up-regulates GADD34, which in turn dephosphorylates eIF2α.